Gene
KWMTBOMO15983
Annotation
PREDICTED:_protein_lethal(2)essential_for_life-like_isoform_X2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.385
Sequence
CDS
ATGCTACAATTAAGTAAACCATCAAACTTTGGGGACGAATTTGTACTTCGTTCTGTTGATTGGCTCGAGAACCTTCCATGGCACGACGGAAATATTGTGAAAACACAAAAAGACAGATTGGAGATCAATTTACTGGTTAAAGATTTTGCCGCTGAACAAATTCAGGTAAAGGTCGCCGATGATTTTATTGTGATTGAAGCGAAACACGAAGAAAAGAAAGACGAACAGGGATTCATATCTAGACACTTTGTGAGAAAATATAAGTTGCCTCAGGGAAGTCAGCCCAACAAAATCACATCAACGCTATCGGCTGATGGTGTGTTGAAAATAGTGGTGCCAAAAGAAACGGCTGTTTTGAAAGATACTGCGATACCTGTGGCATACGAGGACTCAAAGAAGCCAAATTCGAAATTATGA
Protein
MLQLSKPSNFGDEFVLRSVDWLENLPWHDGNIVKTQKDRLEINLLVKDFAAEQIQVKVADDFIVIEAKHEEKKDEQGFISRHFVRKYKLPQGSQPNKITSTLSADGVLKIVVPKETAVLKDTAIPVAYEDSKKPNSKL
Summary
Similarity
Belongs to the small heat shock protein (HSP20) family.
Uniprot
A0A2H4G8P1
S4PFH3
A0A0L7LSA1
A0A212FJC2
A0A0N0PED4
A0A194QB46
+ More
I3QQD3 A0A2H1WQE6 A0A2D1CRR3 A0A2A4JH73 A0A385PHG6 H9J540 U6BMM6 A0A1E1W5U5 Q2WG64 A0A0A7BZ70 A0A212ERQ7 A0A212F0A4 A0A3S2L9M7 A0A220W0C8 Q1AMF5 A0A222NUL2 A0A2H4LHL7 E2F399 A0A0N1PH36 A0A2H1WKW0 D6WHH1 F1C928 A0A0K0XR50 A0A2W1BH28 Q9GSB6 E2F397 A0A2A4K9P8 A0A2H1W1Y8 E3UKK8 B9VTS8 A0A2A4J9J4 A0A0A7BZF0 A0A0K0XQY0 A0A194Q5N9 A0A0K0XQX1 A0A0A7BZ27 A0A0C5PKY8 Q9GN07 F1C930 A0A194Q6X9 A0A0N1PFQ5 A0A084VQX9 A0A212FIM4 A0A0P0CJ81 R9S0D1 A0A2H1W2W7 S4P142 Q1HAT5 A0A0A7BZM3 A0A2W1BHM1 R9RZX9 Q2LCS7 A0A084VQX7 A0A084VQX8 A0A343EU23 A0A0A7RE39 A0A0N1INR0 A0A182GSM1 A0A023EJ46 Q0KKB1 E2F3A1 A0A182J5R7 A0A2A4J3F8 A0A2H1X0S0 A0A0K0XR51 A0A0L7L8L1 A0A182Q6Z7 Q16JG6 A0A222NUK4 A0A3Q3MJF8 A0A0A7RCE5 Q5R1P7 A0A212FBC6 A0A3S2PJT0 A0A0A7RCH5 R4HL93 A0A2H4G806 Q5R1P4 A0A023EJ33 A0A182G9B2 A0A182G9B4 A0A023EL90 A0A2M4ANC4 A0A182Y117 A0A182J2X5 A0A023ELL3 A0A194QKZ3
I3QQD3 A0A2H1WQE6 A0A2D1CRR3 A0A2A4JH73 A0A385PHG6 H9J540 U6BMM6 A0A1E1W5U5 Q2WG64 A0A0A7BZ70 A0A212ERQ7 A0A212F0A4 A0A3S2L9M7 A0A220W0C8 Q1AMF5 A0A222NUL2 A0A2H4LHL7 E2F399 A0A0N1PH36 A0A2H1WKW0 D6WHH1 F1C928 A0A0K0XR50 A0A2W1BH28 Q9GSB6 E2F397 A0A2A4K9P8 A0A2H1W1Y8 E3UKK8 B9VTS8 A0A2A4J9J4 A0A0A7BZF0 A0A0K0XQY0 A0A194Q5N9 A0A0K0XQX1 A0A0A7BZ27 A0A0C5PKY8 Q9GN07 F1C930 A0A194Q6X9 A0A0N1PFQ5 A0A084VQX9 A0A212FIM4 A0A0P0CJ81 R9S0D1 A0A2H1W2W7 S4P142 Q1HAT5 A0A0A7BZM3 A0A2W1BHM1 R9RZX9 Q2LCS7 A0A084VQX7 A0A084VQX8 A0A343EU23 A0A0A7RE39 A0A0N1INR0 A0A182GSM1 A0A023EJ46 Q0KKB1 E2F3A1 A0A182J5R7 A0A2A4J3F8 A0A2H1X0S0 A0A0K0XR51 A0A0L7L8L1 A0A182Q6Z7 Q16JG6 A0A222NUK4 A0A3Q3MJF8 A0A0A7RCE5 Q5R1P7 A0A212FBC6 A0A3S2PJT0 A0A0A7RCH5 R4HL93 A0A2H4G806 Q5R1P4 A0A023EJ33 A0A182G9B2 A0A182G9B4 A0A023EL90 A0A2M4ANC4 A0A182Y117 A0A182J2X5 A0A023ELL3 A0A194QKZ3
Pubmed
EMBL
KU522477
APX61063.1
GAIX01004007
JAA88553.1
JTDY01000199
KOB78327.1
+ More
AGBW02008280 OWR53835.1 OWR53836.1 KQ459890 KPJ19556.1 KQ459460 KPJ00646.1 JQ707904 AFK14098.1 ODYU01010279 SOQ55273.1 KX977581 ATN45240.1 NWSH01001391 PCG71425.1 MG816464 AYA93248.1 BABH01042227 KF471527 AHA36865.1 GDQN01008714 JAT82340.1 AB214974 BAE48744.1 KJ461913 AHW45915.1 AGBW02012988 OWR44131.1 AGBW02011158 OWR47131.1 RSAL01000078 RVE48718.1 KY701309 ASK86231.1 DQ063591 AAZ14792.1 KX958479 ASQ43195.1 KX845562 KZ150055 ATB54993.1 PZC74337.1 HM046615 ADK55522.1 KQ460141 KPJ17473.1 ODYU01009326 SOQ53668.1 KQ971321 EFA00665.1 HQ219475 ADX96000.1 KP843903 AKS40082.1 KZ150133 PZC73025.1 BABH01025846 AF315318 AAG30945.2 HM046613 ADK55520.1 NWSH01000008 PCG80951.1 ODYU01005718 SOQ46842.1 HM449882 ADO33017.1 FJ602788 ACM24354.1 NWSH01002265 PCG68755.1 KJ461916 AHW45918.1 KP843906 AKS40085.1 KPJ00669.1 KP843896 AKS40075.1 KJ461920 AHW45922.1 KM881570 AJQ22645.1 AF315317 AF315319 FJ602772 AAG30944.1 AAG30946.1 ACM24338.1 HQ219477 ADX96002.1 KPJ00755.1 KQ460891 KPJ10838.1 ATLV01015347 KE525007 KFB40373.1 AGBW02008361 OWR53574.1 KR137569 ALI87024.1 KC710018 KC710022 AGM90551.1 AGM90555.1 SOQ46844.1 GAIX01009066 JAA83494.1 JX491641 KC710020 AB244517 AGC23337.1 AGM90553.1 BAE94664.1 KJ461917 AHW45919.1 PZC74338.1 KC710019 KC710024 RSAL01000012 AGM90552.1 AGM90557.1 RVE53491.1 DQ336356 ABC68342.2 ATLV01015346 KFB40371.1 KFB40372.1 MF067296 ASM61894.1 KM217077 KM217078 AJA32865.1 KPJ10837.1 JXUM01084944 KQ563469 KXJ73789.1 GAPW01004647 JAC08951.1 AB251898 BAF03558.1 HM046617 ADK55524.1 AXCP01009179 NWSH01003339 PCG66520.1 PCG66521.1 ODYU01012514 SOQ58867.1 KP843904 AKS40083.1 JTDY01002346 KOB71621.1 AXCN02000983 CH478011 CH477680 EAT34406.1 EAT37335.1 KX958471 ASQ43187.1 KM217080 AJA32868.1 AB195970 BAD74195.1 AGBW02009356 OWR51030.1 RVE53492.1 KM217079 AJA32867.1 HQ647012 AET10424.1 KU522480 APX61066.1 AB195973 BAD74198.1 GAPW01004646 JAC08952.1 JXUM01049171 KQ561594 KXJ78061.1 JXUM01049173 KXJ78063.1 GAPW01004599 JAC08999.1 GGFK01008936 MBW42257.1 GAPW01004454 JAC09144.1 KQ458671 KPJ05575.1
AGBW02008280 OWR53835.1 OWR53836.1 KQ459890 KPJ19556.1 KQ459460 KPJ00646.1 JQ707904 AFK14098.1 ODYU01010279 SOQ55273.1 KX977581 ATN45240.1 NWSH01001391 PCG71425.1 MG816464 AYA93248.1 BABH01042227 KF471527 AHA36865.1 GDQN01008714 JAT82340.1 AB214974 BAE48744.1 KJ461913 AHW45915.1 AGBW02012988 OWR44131.1 AGBW02011158 OWR47131.1 RSAL01000078 RVE48718.1 KY701309 ASK86231.1 DQ063591 AAZ14792.1 KX958479 ASQ43195.1 KX845562 KZ150055 ATB54993.1 PZC74337.1 HM046615 ADK55522.1 KQ460141 KPJ17473.1 ODYU01009326 SOQ53668.1 KQ971321 EFA00665.1 HQ219475 ADX96000.1 KP843903 AKS40082.1 KZ150133 PZC73025.1 BABH01025846 AF315318 AAG30945.2 HM046613 ADK55520.1 NWSH01000008 PCG80951.1 ODYU01005718 SOQ46842.1 HM449882 ADO33017.1 FJ602788 ACM24354.1 NWSH01002265 PCG68755.1 KJ461916 AHW45918.1 KP843906 AKS40085.1 KPJ00669.1 KP843896 AKS40075.1 KJ461920 AHW45922.1 KM881570 AJQ22645.1 AF315317 AF315319 FJ602772 AAG30944.1 AAG30946.1 ACM24338.1 HQ219477 ADX96002.1 KPJ00755.1 KQ460891 KPJ10838.1 ATLV01015347 KE525007 KFB40373.1 AGBW02008361 OWR53574.1 KR137569 ALI87024.1 KC710018 KC710022 AGM90551.1 AGM90555.1 SOQ46844.1 GAIX01009066 JAA83494.1 JX491641 KC710020 AB244517 AGC23337.1 AGM90553.1 BAE94664.1 KJ461917 AHW45919.1 PZC74338.1 KC710019 KC710024 RSAL01000012 AGM90552.1 AGM90557.1 RVE53491.1 DQ336356 ABC68342.2 ATLV01015346 KFB40371.1 KFB40372.1 MF067296 ASM61894.1 KM217077 KM217078 AJA32865.1 KPJ10837.1 JXUM01084944 KQ563469 KXJ73789.1 GAPW01004647 JAC08951.1 AB251898 BAF03558.1 HM046617 ADK55524.1 AXCP01009179 NWSH01003339 PCG66520.1 PCG66521.1 ODYU01012514 SOQ58867.1 KP843904 AKS40083.1 JTDY01002346 KOB71621.1 AXCN02000983 CH478011 CH477680 EAT34406.1 EAT37335.1 KX958471 ASQ43187.1 KM217080 AJA32868.1 AB195970 BAD74195.1 AGBW02009356 OWR51030.1 RVE53492.1 KM217079 AJA32867.1 HQ647012 AET10424.1 KU522480 APX61066.1 AB195973 BAD74198.1 GAPW01004646 JAC08952.1 JXUM01049171 KQ561594 KXJ78061.1 JXUM01049173 KXJ78063.1 GAPW01004599 JAC08999.1 GGFK01008936 MBW42257.1 GAPW01004454 JAC09144.1 KQ458671 KPJ05575.1
Proteomes
Pfam
PF00011 HSP20
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2H4G8P1
S4PFH3
A0A0L7LSA1
A0A212FJC2
A0A0N0PED4
A0A194QB46
+ More
I3QQD3 A0A2H1WQE6 A0A2D1CRR3 A0A2A4JH73 A0A385PHG6 H9J540 U6BMM6 A0A1E1W5U5 Q2WG64 A0A0A7BZ70 A0A212ERQ7 A0A212F0A4 A0A3S2L9M7 A0A220W0C8 Q1AMF5 A0A222NUL2 A0A2H4LHL7 E2F399 A0A0N1PH36 A0A2H1WKW0 D6WHH1 F1C928 A0A0K0XR50 A0A2W1BH28 Q9GSB6 E2F397 A0A2A4K9P8 A0A2H1W1Y8 E3UKK8 B9VTS8 A0A2A4J9J4 A0A0A7BZF0 A0A0K0XQY0 A0A194Q5N9 A0A0K0XQX1 A0A0A7BZ27 A0A0C5PKY8 Q9GN07 F1C930 A0A194Q6X9 A0A0N1PFQ5 A0A084VQX9 A0A212FIM4 A0A0P0CJ81 R9S0D1 A0A2H1W2W7 S4P142 Q1HAT5 A0A0A7BZM3 A0A2W1BHM1 R9RZX9 Q2LCS7 A0A084VQX7 A0A084VQX8 A0A343EU23 A0A0A7RE39 A0A0N1INR0 A0A182GSM1 A0A023EJ46 Q0KKB1 E2F3A1 A0A182J5R7 A0A2A4J3F8 A0A2H1X0S0 A0A0K0XR51 A0A0L7L8L1 A0A182Q6Z7 Q16JG6 A0A222NUK4 A0A3Q3MJF8 A0A0A7RCE5 Q5R1P7 A0A212FBC6 A0A3S2PJT0 A0A0A7RCH5 R4HL93 A0A2H4G806 Q5R1P4 A0A023EJ33 A0A182G9B2 A0A182G9B4 A0A023EL90 A0A2M4ANC4 A0A182Y117 A0A182J2X5 A0A023ELL3 A0A194QKZ3
I3QQD3 A0A2H1WQE6 A0A2D1CRR3 A0A2A4JH73 A0A385PHG6 H9J540 U6BMM6 A0A1E1W5U5 Q2WG64 A0A0A7BZ70 A0A212ERQ7 A0A212F0A4 A0A3S2L9M7 A0A220W0C8 Q1AMF5 A0A222NUL2 A0A2H4LHL7 E2F399 A0A0N1PH36 A0A2H1WKW0 D6WHH1 F1C928 A0A0K0XR50 A0A2W1BH28 Q9GSB6 E2F397 A0A2A4K9P8 A0A2H1W1Y8 E3UKK8 B9VTS8 A0A2A4J9J4 A0A0A7BZF0 A0A0K0XQY0 A0A194Q5N9 A0A0K0XQX1 A0A0A7BZ27 A0A0C5PKY8 Q9GN07 F1C930 A0A194Q6X9 A0A0N1PFQ5 A0A084VQX9 A0A212FIM4 A0A0P0CJ81 R9S0D1 A0A2H1W2W7 S4P142 Q1HAT5 A0A0A7BZM3 A0A2W1BHM1 R9RZX9 Q2LCS7 A0A084VQX7 A0A084VQX8 A0A343EU23 A0A0A7RE39 A0A0N1INR0 A0A182GSM1 A0A023EJ46 Q0KKB1 E2F3A1 A0A182J5R7 A0A2A4J3F8 A0A2H1X0S0 A0A0K0XR51 A0A0L7L8L1 A0A182Q6Z7 Q16JG6 A0A222NUK4 A0A3Q3MJF8 A0A0A7RCE5 Q5R1P7 A0A212FBC6 A0A3S2PJT0 A0A0A7RCH5 R4HL93 A0A2H4G806 Q5R1P4 A0A023EJ33 A0A182G9B2 A0A182G9B4 A0A023EL90 A0A2M4ANC4 A0A182Y117 A0A182J2X5 A0A023ELL3 A0A194QKZ3
PDB
3J07
E-value=3.6659e-20,
Score=235
Ontologies
GO
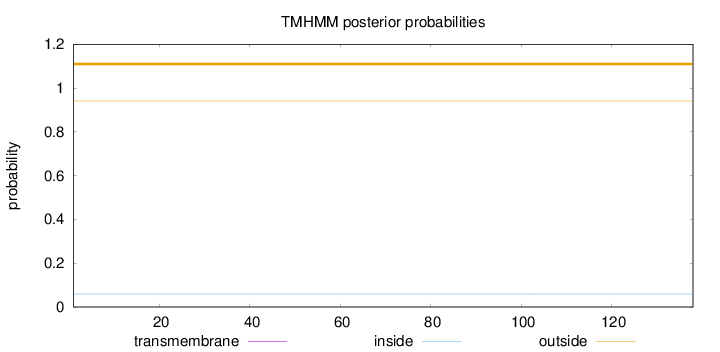
Topology
Length:
138
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05909
outside
1 - 138
Population Genetic Test Statistics
Pi
195.933501
Theta
172.893363
Tajima's D
0.40956
CLR
0
CSRT
0.485875706214689
Interpretation
Uncertain
