Gene
KWMTBOMO15970
Pre Gene Modal
BGIBMGA005714
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_latrophilin_Cirl-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.139 PlasmaMembrane Reliability : 1.949
Sequence
CDS
ATGAGCTCAGAGCCACGATATGAGACAGCGTACGCCTGTGAAGGGAAGACCCTTAAGATAGGCTGCGGCGAGGGATCCGTGATTCATCTAATAAGGGCGAACTACGGTCGTTTCTCGATAACGATATGCAATGACCACGGGAACACCGACTGGAGCGTAAATTGCATGTCCACGAGGAGTTTGAGGGTTTTGCATAGCAAATGCAGCATGCACCAGAATTGCAGTATATCGGCCAACACGAATATATTTGGTGATCCGTGTCCTAATACGTTGAAGTATTTAGAAGCGCATTATCAATGTGTGCCGGCTTCAACAACAAGCACGACCAACAGACCGTCTCCTCCCTGGCTAATCACGTCTCCTCAAACGGTGTGGAGGTCCACCATCGCTCCGCTACCGAAGAACCCAGTCCAAACGCAGAATGAGAGGAAGAAACCAACTGTGATCACACCGCCCACATTCAGGCCTCACATTACTTTACCTCCAGAAGTAAAGTTGGGCGACATTGGGACCAAGAAGACAACTCTAAGACAACCAGATACATCGTCGGAGAAGGAGCCTTCAGTGAGCCCCAAGGACGTCATTCCTGAAGAGAAAGATGAAACACCCAAAGAAGTTGAACTAGACCAAAACCAAAGAAACGACAATCCTGCGATAGAAGAAGATCCAGAACCAGTAACTGAAGAACCACCTAAAGAAGATGATTCCGATTTGACTGTATCAGTCAATTCACCACCAAAAGACAAACATGATATTCTGCAAGATTTATTCCCAACGAAAGAAATAAAAGCAATCGCCCGCCACGCTTACTGTCCTCAAGTGACAGCTAGAGGCTTACTGTGGAATTGGACATTGGCGGAGACTTATGGGGTGCAGCCGTGTCCTGGTGGAGCTACCGGACTGGCTCGATGGAAATGCACACTAACACCACATTACTACGTTGGAGACGAAAAACCAAGAAGACGAACGGGTCTACCCTCAGGAGAGAATTTCCATGATTTAGGTCTAGACCAACCCGTGGACAAGCACTTGCAATTGGTCCGACGGCTGGGCTCGGAAGTGCTCCTTAAAGGCCGCAATGGTGACACGCCAGAACTAATGGTCGATTATAGAGGGAGCAGCCTTCCTGATGCTCAGGCATCTAGTGACAACTCTCTCATCAATGTTGAACAGATTTACACAGATTTAGTACCAGAATGGCATGGAGCATCACCAGACCTTAGCGAGTGCAGATCCGTATGGCTGAACAATTTGGAAGCGAGGGTCAGAGAAGGAGAATCGCTACTTAGTATCAGCAATGATTTGGCACAGGTAACATCTTCGAAGACCCTCTACGGCGGAGACATGATGACAACTACGAGTATCATGAAGAAGCTAGCCCAGCGGATGTCGCAGAACATACAAAGCTTCCGGGACCCGAGCCAAAGAGAAGCTCTCGTTACCGATATGCTGCTTGGAGTTATTAAAACTGGATCGAATCTTCTAGAAGAGAGTCAGCGAGCGTCCTGGGGCGACTTGAGCGTGGAAGGACAGAACCGCGTCGCCAGTGCACTGCTCACGCACTTAGAAGAAAACGCCTTCTTACTAGCAGATGCCGTTTCAAAAGAGAAAACAATTTTACAAATCGTCAAAAACATATGCCTCTCCGTGAGAGTTTTGGAAGTGAAAAACGTAGAAGATGAAACATTCCCATCTGTGATAGCCCAAGAACAGTGGAAGGCTTCAGAAGACATCATTACGATACCAAAAGCTGCTTTATTAGACAACCGACAAAACGATCTGGTCAGGATTGTATTCTTCGCGTTCGATCGCCTCGAAGAAATTCTGCCCCCGACCTTCGGCAAGACCGCCCCTCACTACGCGCCCCACCGCTCCGAGGTCGGGGGGGCGCCCGCGTCCTCCACCAGCATTAACTCTGAGAAGGAAAAGAAAAATATCACAAGGCTACTGAATTCCAAGGTGATATCCGCGAGTTTAGGGAACGGCCGCCATATCCAGCTCTCGCAAATGGTCAAAGTCACCATGAAGCATCTGCAAACGGAGAACGTAACCAATCCCGCCTGCGTGTTCTGGGATTATACTTTGCATGGCTGGTCCCCTGAAGGTTGTGAGGTGGAGTGGACGAACTCGACTCACACCGGCTGCGCGTGCTCCCACCTCACGAACTTCGCCGTCCTGATGGACGTTCACGGCGTGGCCCTCAGCGCTGCCCACGAGCTCGCCCTGCAGACCCTGACGTACGTCGGCTGCGGGATATCCACTGTCGCTCTGTCTGCTGCCATGATCGTGTTCACCTGCTTCGGGAACACTAAGTCGGATCGCAGCTTAATCCATAAGAACCTATGCGCCTGTCTGCTTGTCGCCGAAGTAGTGTTTCTGGCCGGTATCGATAAAACACAGAATAGAATATTCTGCGGGATCATCGCTGGGCTGTTGCACTATTTCTTCTTGGCGGCCTTCGCTTGGATGTTTCTAGAAGGGTTCCACCTGTACGCGATGCTGGTGGAGGTGTTCGAGCCGGAGCACCCTCGCGCGCGCTGGTACTGCGCATGCGCGTACCTGGCGCCGGCGCTGGTGGTGCTCGCGTCGGCGGCCGCGTACCCCGGCGGCTACGGCACCGCCCAGCACTGCTGGCTCTCCACCGACCGCATGTTCATCATGAGCTTCGTCGGACCCGTCACCTTGGTTTTGTTGGCCAACTTGATATGTCTGAGCATGGTAATCTACATGATGTGCACTCACTCCACTGTGTCGCTGAAGGCCAAAGACGATTCCAAGCTATACAAGATCAAGGTCTGGTTGAAGAGCTCCCTCGTTTTAGTGTTCTTGTTGGGCTTGACTTGGGCTTTCGGTCTTCTCTACCTAAACCAGCAGACAGTGGTCATGGCTTATGCGTTTACCATCCTGAACTCCCTCCAAGGGTTGGTCATCTTCATATTCCATTGTCTCCAGAATGACAAGTTCCGCAACGAATGCGCTCGTCTGAGTCGTCGCCACCGCTGGCTCTCGTGCTGCTTGTACTGGACGGGGCAGCCCTCGGAGACCCTTCAGCACAACAATGGACCCTCGGGGTCCAGCTCGAACCACGTCGACACGCGCCAGGGCTCCGTCAAGAGCAGGAGGTACCACCAGGCAGCGCCGCCTGACGACGAATTGGACCCCAGAGATGTGACGAACACTTCTTCTAAACACCACGTTGGGAACACTTCAACGCTACCGCATCACGACACTCTCTCCAGGCAACTGGCGCCCCCAAATCATCTCCACAGCCCTTACGAGTGCGAACCGGTGGCTCCTGGCTCACCGCGCCACATGAAAGACGAAGAAAGCCCCCTCCATCATCAAGGATACCAGACCACGCGGAGCTACAACCACGACTTCCGGAACCCCCACGTCTACATGTCCCAGCACTCGCCGGGAAGTCAAGAGATGATTTTTCGGAAGCATTACAAGGACGACCGCAGGAAACACCACCATCACCACGGAAGCCAGAACATGAACCACACGTATTCGGAGATAGCGTCCCAACAGGGCCGGATGTACAGGCGCGGATCCGAACAGGAGTTCCGCGTGATCCAGGACGATCCGGTTTACGAGGAGATAGAGAGAAACGAAACGATGATGTCAGACGTGTCCGACGACAACTCCGGACCGGACTGCAGACAGAATGTAGGTCACCACGGCTCATCTAGCTCCAAGTTTTTTGGGGACCACCGACCGCTGATATCGTACAGCCCAGGCGAGAGGCACCACCACGACCCGGAGCATTACGGGAAGTACGGTAAATGGGACACGTTAGAGCGCTACGATCCGAGGCATTACGAAAGCGGTCGGCTGATGCACCAATACCACCAGCCTCAAGAGAACATGAGGGCGCTTGCGGCCGTCCTCAACGGGGAGAACAACGTCGTCTGCCATTTGGAGCCTCACAATTCTTACGACGTGTACCAGCCTCAGAGCGGCAACCCCAGGACTGTATCCCAGCCCAGTTTCTGA
Protein
MSSEPRYETAYACEGKTLKIGCGEGSVIHLIRANYGRFSITICNDHGNTDWSVNCMSTRSLRVLHSKCSMHQNCSISANTNIFGDPCPNTLKYLEAHYQCVPASTTSTTNRPSPPWLITSPQTVWRSTIAPLPKNPVQTQNERKKPTVITPPTFRPHITLPPEVKLGDIGTKKTTLRQPDTSSEKEPSVSPKDVIPEEKDETPKEVELDQNQRNDNPAIEEDPEPVTEEPPKEDDSDLTVSVNSPPKDKHDILQDLFPTKEIKAIARHAYCPQVTARGLLWNWTLAETYGVQPCPGGATGLARWKCTLTPHYYVGDEKPRRRTGLPSGENFHDLGLDQPVDKHLQLVRRLGSEVLLKGRNGDTPELMVDYRGSSLPDAQASSDNSLINVEQIYTDLVPEWHGASPDLSECRSVWLNNLEARVREGESLLSISNDLAQVTSSKTLYGGDMMTTTSIMKKLAQRMSQNIQSFRDPSQREALVTDMLLGVIKTGSNLLEESQRASWGDLSVEGQNRVASALLTHLEENAFLLADAVSKEKTILQIVKNICLSVRVLEVKNVEDETFPSVIAQEQWKASEDIITIPKAALLDNRQNDLVRIVFFAFDRLEEILPPTFGKTAPHYAPHRSEVGGAPASSTSINSEKEKKNITRLLNSKVISASLGNGRHIQLSQMVKVTMKHLQTENVTNPACVFWDYTLHGWSPEGCEVEWTNSTHTGCACSHLTNFAVLMDVHGVALSAAHELALQTLTYVGCGISTVALSAAMIVFTCFGNTKSDRSLIHKNLCACLLVAEVVFLAGIDKTQNRIFCGIIAGLLHYFFLAAFAWMFLEGFHLYAMLVEVFEPEHPRARWYCACAYLAPALVVLASAAAYPGGYGTAQHCWLSTDRMFIMSFVGPVTLVLLANLICLSMVIYMMCTHSTVSLKAKDDSKLYKIKVWLKSSLVLVFLLGLTWAFGLLYLNQQTVVMAYAFTILNSLQGLVIFIFHCLQNDKFRNECARLSRRHRWLSCCLYWTGQPSETLQHNNGPSGSSSNHVDTRQGSVKSRRYHQAAPPDDELDPRDVTNTSSKHHVGNTSTLPHHDTLSRQLAPPNHLHSPYECEPVAPGSPRHMKDEESPLHHQGYQTTRSYNHDFRNPHVYMSQHSPGSQEMIFRKHYKDDRRKHHHHHGSQNMNHTYSEIASQQGRMYRRGSEQEFRVIQDDPVYEEIERNETMMSDVSDDNSGPDCRQNVGHHGSSSSKFFGDHRPLISYSPGERHHHDPEHYGKYGKWDTLERYDPRHYESGRLMHQYHQPQENMRALAAVLNGENNVVCHLEPHNSYDVYQPQSGNPRTVSQPSF
Summary
Similarity
Belongs to the G-protein coupled receptor 2 family.
Uniprot
EMBL
Proteomes
Interpro
IPR000922
Lectin_gal-bd_dom
+ More
IPR000832 GPCR_2_secretin-like
IPR036445 GPCR_2_extracell_dom_sf
IPR031240 Latrophilin-2
IPR017981 GPCR_2-like
IPR000203 GPS
IPR032471 GAIN_dom_N
IPR031234 Latrophilin-1
IPR017983 GPCR_2_secretin-like_CS
IPR008077 GPCR_2_brain_angio_inhib
IPR015630 Latrophilin-3
IPR001879 GPCR_2_extracellular_dom
IPR042099 AMP-dep_Synthh-like_sf
IPR000873 AMP-dep_Synth/Lig
IPR000832 GPCR_2_secretin-like
IPR036445 GPCR_2_extracell_dom_sf
IPR031240 Latrophilin-2
IPR017981 GPCR_2-like
IPR000203 GPS
IPR032471 GAIN_dom_N
IPR031234 Latrophilin-1
IPR017983 GPCR_2_secretin-like_CS
IPR008077 GPCR_2_brain_angio_inhib
IPR015630 Latrophilin-3
IPR001879 GPCR_2_extracellular_dom
IPR042099 AMP-dep_Synthh-like_sf
IPR000873 AMP-dep_Synth/Lig
Gene 3D
ProteinModelPortal
PDB
4DLQ
E-value=8.72495e-34,
Score=364
Ontologies
GO
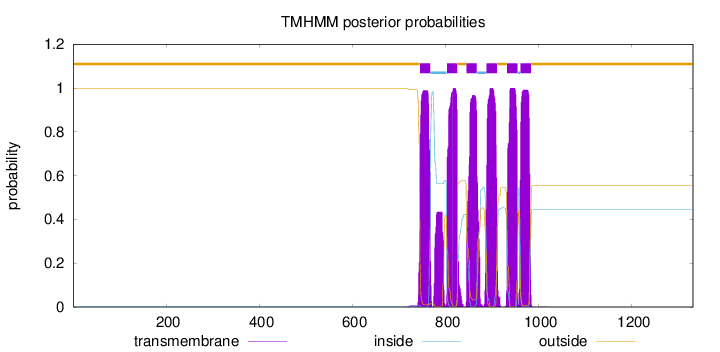
Topology
Length:
1331
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
142.24993
Exp number, first 60 AAs:
0.00097
Total prob of N-in:
0.00143
outside
1 - 744
TMhelix
745 - 767
inside
768 - 802
TMhelix
803 - 825
outside
826 - 844
TMhelix
845 - 867
inside
868 - 887
TMhelix
888 - 910
outside
911 - 931
TMhelix
932 - 954
inside
955 - 960
TMhelix
961 - 983
outside
984 - 1331
Population Genetic Test Statistics
Pi
217.435472
Theta
148.464
Tajima's D
1.433726
CLR
0.667517
CSRT
0.764411779411029
Interpretation
Uncertain
