Gene
KWMTBOMO15964
Pre Gene Modal
BGIBMGA005722
Annotation
PREDICTED:_endothelin-converting_enzyme_1_[Bombyx_mori]
Full name
Neprilysin-3
+ More
Endothelin-converting enzyme homolog
Endothelin-converting enzyme homolog
Location in the cell
Cytoplasmic Reliability : 1.346
Sequence
CDS
ATGCCAACTCTACATCTGAAACCCGGGATCACGATGGCTCGTTACAAGAAGACTGGTTTTATGGACGACGATGCCGGAAGCATCAGCTCGATCCAGCTCAACGAAGGAGTACGGTCCGGAGGAACTCATATTCGCTATCATATGGGTGTATCGTTCATAGGATGGCATTCGCGTACAAGATTGGAGCGTGTTCTGCTCGTGACCAGCGCTACGCTCTTTGTGGGTGTACTGCTCATGGTGACTCTTTTGCTAACCACAAGAGGTACTCCGGATATCCCTGTGCCGCCATGTTATCAGCCTGAACCTAAAGGCCCAAGCATTTGTCTTTCGAGCTCCTGCATTTACACTGCCAGCGAAGTTATCCAGGCCCTGGACGAAACTAAGGACCCGTGCGATGACTTCTACGAGTTCGCGTGTGGAGGATGGCTGAGGAACAATCCAATTCCTGAAGGAAAATCTACATGGGGTATCTTTAGTAAGATCGAGCTGCAGAACCAGTTGATCATCAGGTCCGCTCTGGAAAAGGTGAATACTACAAACAAATCGGAAGCAGAATATAAGGCGCGCGTGTATTATGATGCGTGCATCGACACGAATGAAACAATAGAGAAACTGGCGGAGAAACCTTTGATAGCCGTCATCAAGAAACTCGGAGGCTGGCATATACTGCCGTCAACGATGGTCAAACAGCAGAAATGGGACTTCCAGAAGCTGGTTCAGGACGTACAGAACTCATACAACCTGGGAGGCCTCTTCAACTGGGCCGTGTCTGAGGACGACAGAAACTCATCCAAACACGTCATCGTTCTTGACCAAGGAGGTCTTAATCTACCAACACGAGACAACTACCTGAACAAAACCGCCCATAAGAAGGTCCTAGACGCATATCTAGATTTTATGACTAGGATCTGTGTCCTCTTAGGCGCAAACAAATCCGAGGCTCGTGCCCAAATGAGCAAGGTCATTCAGTTCGAAACAGATTTAGCTAATATTACTACGCCCTCTGAAGACAGAAGAGATGAAGAAGGAATGTATAATCCTTATACAATAAGGCAGTGGCAGAAGGAAGTGTCTTTCTTGAATTGGACCCAGTTTTTTAATGATGCATTTAAGCTTGTCAATCGTACTATTCCAGATACGCAGAGAATTGTCGTTTACGCGCCAGACTACTTCAAGAATTTGACGAGGATCATACAAAAGTACAATAAAACTGAAGAAGACCAGAGAACGATAACCAGTTACATGATGTGGCAAGTTTCAAGATCTCTCTCCACTTATTTATCGAAGCCTTTTCGGGACGCCACCAAGATTCTTAGGAAGGCGCTTTTCGGATCTGAAGGCACCGAAGAGTCTTGGAGATACTGCGTCACTGACACTAACAATGCTATTGGTTTCGCGGTTGGAGCCATGTTCGTCCGCGAAGTCTTCCACGGCGAAGCGAAGACCCAGGGAGAAATAATGATTGATAACATCCGTAACGCTTTCAAGGAGAACCTGAGGAATTTGGACTGGATGGACACCGAGACGAGGAAAGCGGCCGAGACGAAGGCTGACGCTATAACTGATATGATCGGTTTCCCGGACTACATTCTCCACAAAGACGAACTGAACAAGCAATATGAAGAGTTGGTCGTTAAGCCAAACGAATACTTCGAAAATAACATAGCTTACAACATGTATTCGCTTAAGAACGACCTATTAAAACTGGATAAACCTGTCAACAAATCAAAATGGGGAATGACTCCGTCAACGGTAAACGCTTACTACACTCCCACGAAGAACCAAATAGTGTTTCCGGCCGGGATCCTGCAGTTGCCTTTCTATGATGGGGACAACCCGAAGTGCGTGAACTACGGGGCCATGGGGGTCGTCATGGGACACGAGCTGACCCACGCCTTTGACGATCAAGGAAGAGAATACGACAGATTTGGCAACTTGAACCAGTGGTGGAACGACGCGACGATTGCGAAATTCAAAAAACGCGCCGAATGCATTCGAGAACAATATTCGAAATATACAATGGACGGACATCATCTAAACGGGAAACAAACATTAGGGGAGAACATAGCCGACAACGGTGGCCTCAAAGCGTCATTCCACGCTTACCTGGACTACACCAAGAAAGCATCAGTTAACCTAACACTGCCGGGACTGAAATACAACGATAGACAACTTTTCTTTATTTCTTTTGCACAGGTATGGTGCTCGGCAATGACTAAAGAATCAACGAAAATGCAAATTGAGAAGGACGATCATACAGTTGCCAAATATCGAGTTATAGGACCAATAGCAAACCTAAAGGAGTTCTCCGACGAATTTCACTGTCCAATTGGTTCGAAAATGAATCCAACACATAAATGTGAAGTTTGGTAA
Protein
MPTLHLKPGITMARYKKTGFMDDDAGSISSIQLNEGVRSGGTHIRYHMGVSFIGWHSRTRLERVLLVTSATLFVGVLLMVTLLLTTRGTPDIPVPPCYQPEPKGPSICLSSSCIYTASEVIQALDETKDPCDDFYEFACGGWLRNNPIPEGKSTWGIFSKIELQNQLIIRSALEKVNTTNKSEAEYKARVYYDACIDTNETIEKLAEKPLIAVIKKLGGWHILPSTMVKQQKWDFQKLVQDVQNSYNLGGLFNWAVSEDDRNSSKHVIVLDQGGLNLPTRDNYLNKTAHKKVLDAYLDFMTRICVLLGANKSEARAQMSKVIQFETDLANITTPSEDRRDEEGMYNPYTIRQWQKEVSFLNWTQFFNDAFKLVNRTIPDTQRIVVYAPDYFKNLTRIIQKYNKTEEDQRTITSYMMWQVSRSLSTYLSKPFRDATKILRKALFGSEGTEESWRYCVTDTNNAIGFAVGAMFVREVFHGEAKTQGEIMIDNIRNAFKENLRNLDWMDTETRKAAETKADAITDMIGFPDYILHKDELNKQYEELVVKPNEYFENNIAYNMYSLKNDLLKLDKPVNKSKWGMTPSTVNAYYTPTKNQIVFPAGILQLPFYDGDNPKCVNYGAMGVVMGHELTHAFDDQGREYDRFGNLNQWWNDATIAKFKKRAECIREQYSKYTMDGHHLNGKQTLGENIADNGGLKASFHAYLDYTKKASVNLTLPGLKYNDRQLFFISFAQVWCSAMTKESTKMQIEKDDHTVAKYRVIGPIANLKEFSDEFHCPIGSKMNPTHKCEVW
Summary
Description
Metalloendoprotease which is required in the dorsal paired medial neurons for the proper formation of long-term (LTM) and middle-term memories (MTM). Also required in the mushroom body neurons where it functions redundantly with neprilysins Nep2 and Nep4 in normal LTM formation.
Catalytic Activity
Preferential cleavage of polypeptides between hydrophobic residues, particularly with Phe or Tyr at P1'.
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M13 family.
Keywords
Cell membrane
Complete proteome
Disulfide bond
Glycoprotein
Hydrolase
Membrane
Metal-binding
Metalloprotease
Protease
Reference proteome
Signal-anchor
Transmembrane
Transmembrane helix
Zinc
Feature
chain Neprilysin-3
Uniprot
H9J880
A0A3S2M7Y4
A0A194PWR1
A0A2A4JPV3
A0A2H1WST7
A0A212EJ44
+ More
A0A182QD14 A0A182XZM3 A0A182K4Z9 A0A182T7L3 A0A182MZC1 A0A1Y9IUX2 A0A182UXA3 A0A1S4GX10 A0A182P0D4 A0A182TST7 A0A1Y9IVL3 A0A182X1I7 A0A182RU97 Q7Q3B4 A0A182L2I9 A0A182IEG8 A0A1Y9IXE8 A0A182MER4 A0A084VI29 A0A240PLB3 A0A182J972 A0A2J7RDW6 A0A0C9S1N5 A0A1Q3G4C0 K7IVN1 A0A1Q3G4E4 A0A0C9RMP5 A0A1Q3G4D3 A0A1Q3G4A4 A0A1Q3G4C3 A0A067REZ8 A0A232ENX7 W8BGE7 Q16Q77 A0A1S4FSY8 W8C2N2 W5J4G0 A0A3L8E1J0 A0A2M4CG58 A0A026WJ96 A0A2M4CGE5 A0A2M4A6X3 A0A1B6E866 A0A1A9YGV4 A0A1A9UV25 A0A1B6JFG3 A0A1Q3G491 B3MQK6 B4PYY9 A0A0R1EAQ4 A0A0P8Y4K7 A0A1B0FM21 A0A0P8YKQ7 B3NWE8 A0A1B0AJ08 A0A1I8JSH2 Q9W5Y0 A0A1B0AT16 A0A0Q5T447 A0A1W4UQT4 A0A182F1R2 A0A1Q3G498 A0A1W4UR29 Q8IS64 E1ZWD4 A0A2M4BEE5 A0A2M4BED4 A0A0R3P4T4 A0A2A3EQ90 Q29GV1 A0A023F3J7 E2B3X1 A0A0R3P5C3 A0A0R3P0H4 A0A3B0JYR1 B4N1N0 A0A0Q9X3K1 A0A087ZS76 A0A182GDM3 B4JMG9 A0A154PJ26 A0A0L7RCP1 A0A0P4VL55 A0A310SFY3 B4M2F3 A0A0Q9WCR8 A0A0M4F9B9 A0A2M3Z5A3 A0A0N0BJI3 A0A336LY78 A0A1Y1K5Z5 D2A597 A0A1B0CGE7
A0A182QD14 A0A182XZM3 A0A182K4Z9 A0A182T7L3 A0A182MZC1 A0A1Y9IUX2 A0A182UXA3 A0A1S4GX10 A0A182P0D4 A0A182TST7 A0A1Y9IVL3 A0A182X1I7 A0A182RU97 Q7Q3B4 A0A182L2I9 A0A182IEG8 A0A1Y9IXE8 A0A182MER4 A0A084VI29 A0A240PLB3 A0A182J972 A0A2J7RDW6 A0A0C9S1N5 A0A1Q3G4C0 K7IVN1 A0A1Q3G4E4 A0A0C9RMP5 A0A1Q3G4D3 A0A1Q3G4A4 A0A1Q3G4C3 A0A067REZ8 A0A232ENX7 W8BGE7 Q16Q77 A0A1S4FSY8 W8C2N2 W5J4G0 A0A3L8E1J0 A0A2M4CG58 A0A026WJ96 A0A2M4CGE5 A0A2M4A6X3 A0A1B6E866 A0A1A9YGV4 A0A1A9UV25 A0A1B6JFG3 A0A1Q3G491 B3MQK6 B4PYY9 A0A0R1EAQ4 A0A0P8Y4K7 A0A1B0FM21 A0A0P8YKQ7 B3NWE8 A0A1B0AJ08 A0A1I8JSH2 Q9W5Y0 A0A1B0AT16 A0A0Q5T447 A0A1W4UQT4 A0A182F1R2 A0A1Q3G498 A0A1W4UR29 Q8IS64 E1ZWD4 A0A2M4BEE5 A0A2M4BED4 A0A0R3P4T4 A0A2A3EQ90 Q29GV1 A0A023F3J7 E2B3X1 A0A0R3P5C3 A0A0R3P0H4 A0A3B0JYR1 B4N1N0 A0A0Q9X3K1 A0A087ZS76 A0A182GDM3 B4JMG9 A0A154PJ26 A0A0L7RCP1 A0A0P4VL55 A0A310SFY3 B4M2F3 A0A0Q9WCR8 A0A0M4F9B9 A0A2M3Z5A3 A0A0N0BJI3 A0A336LY78 A0A1Y1K5Z5 D2A597 A0A1B0CGE7
EC Number
3.4.24.11
3.4.24.-
3.4.24.-
Pubmed
19121390
26354079
22118469
25244985
12364791
14747013
+ More
17210077 20966253 24438588 20075255 24845553 28648823 24495485 17510324 20920257 23761445 30249741 24508170 17994087 17550304 10731132 12537572 12537569 24395329 27629706 12752656 20798317 15632085 25474469 26483478 27129103 28004739 18362917 19820115
17210077 20966253 24438588 20075255 24845553 28648823 24495485 17510324 20920257 23761445 30249741 24508170 17994087 17550304 10731132 12537572 12537569 24395329 27629706 12752656 20798317 15632085 25474469 26483478 27129103 28004739 18362917 19820115
EMBL
BABH01038191
BABH01038192
BABH01038193
RSAL01000012
RVE53513.1
KQ459590
+ More
KPI97453.1 NWSH01000851 PCG73849.1 ODYU01010405 SOQ55484.1 AGBW02014530 OWR41506.1 AXCN02002075 AAAB01008964 EAA12912.1 APCN01005203 AXCM01004319 ATLV01013267 KE524849 KFB37623.1 NEVH01005280 PNF39029.1 GBYB01014696 JAG84463.1 GFDL01000411 JAV34634.1 GFDL01000406 JAV34639.1 GBYB01014697 JAG84464.1 GFDL01000385 JAV34660.1 GFDL01000415 JAV34630.1 GFDL01000426 JAV34619.1 KK852548 KDR21598.1 NNAY01003046 OXU20060.1 GAMC01010552 JAB96003.1 CH477758 EAT36553.1 GAMC01010551 JAB96004.1 ADMH02002119 ETN58796.1 QOIP01000001 RLU26255.1 GGFL01000152 MBW64330.1 KK107218 EZA55169.1 GGFL01000151 MBW64329.1 GGFK01003057 MBW36378.1 GEDC01022659 GEDC01003191 JAS14639.1 JAS34107.1 GECU01009716 JAS97990.1 GFDL01000413 JAV34632.1 CH902621 EDV44632.1 CM000162 EDX02067.2 KRK06544.1 KPU81656.1 CCAG010015359 KPU81655.1 CH954180 EDV46768.1 AE014298 AY095202 BT132915 AAM12295.1 AEW12887.1 JXJN01003092 JXJN01003093 KQS30114.1 GFDL01000418 JAV34627.1 AY149919 AAN73018.1 GL434809 EFN74503.1 GGFJ01002285 MBW51426.1 GGFJ01002275 MBW51416.1 CH379064 KRT06767.1 KZ288204 PBC33201.1 EAL32008.2 GBBI01002796 JAC15916.1 GL445407 EFN89640.1 KRT06765.1 KRT06766.1 OUUW01000003 SPP78516.1 CH963925 EDW78269.2 KRF98844.1 JXUM01056431 KQ561910 KXJ77198.1 CH916371 EDV91912.1 KQ434931 KZC11845.1 KQ414615 KOC68757.1 GDKW01000749 JAI55846.1 KQ760382 OAD60700.1 CH940651 EDW65857.1 KRF82444.1 CP012528 ALC49112.1 GGFM01002934 MBW23685.1 KQ435714 KOX79211.1 UFQS01000201 UFQT01000201 SSX01253.1 SSX21633.1 GEZM01096133 JAV55135.1 KQ971345 EFA05107.2 AJWK01011058
KPI97453.1 NWSH01000851 PCG73849.1 ODYU01010405 SOQ55484.1 AGBW02014530 OWR41506.1 AXCN02002075 AAAB01008964 EAA12912.1 APCN01005203 AXCM01004319 ATLV01013267 KE524849 KFB37623.1 NEVH01005280 PNF39029.1 GBYB01014696 JAG84463.1 GFDL01000411 JAV34634.1 GFDL01000406 JAV34639.1 GBYB01014697 JAG84464.1 GFDL01000385 JAV34660.1 GFDL01000415 JAV34630.1 GFDL01000426 JAV34619.1 KK852548 KDR21598.1 NNAY01003046 OXU20060.1 GAMC01010552 JAB96003.1 CH477758 EAT36553.1 GAMC01010551 JAB96004.1 ADMH02002119 ETN58796.1 QOIP01000001 RLU26255.1 GGFL01000152 MBW64330.1 KK107218 EZA55169.1 GGFL01000151 MBW64329.1 GGFK01003057 MBW36378.1 GEDC01022659 GEDC01003191 JAS14639.1 JAS34107.1 GECU01009716 JAS97990.1 GFDL01000413 JAV34632.1 CH902621 EDV44632.1 CM000162 EDX02067.2 KRK06544.1 KPU81656.1 CCAG010015359 KPU81655.1 CH954180 EDV46768.1 AE014298 AY095202 BT132915 AAM12295.1 AEW12887.1 JXJN01003092 JXJN01003093 KQS30114.1 GFDL01000418 JAV34627.1 AY149919 AAN73018.1 GL434809 EFN74503.1 GGFJ01002285 MBW51426.1 GGFJ01002275 MBW51416.1 CH379064 KRT06767.1 KZ288204 PBC33201.1 EAL32008.2 GBBI01002796 JAC15916.1 GL445407 EFN89640.1 KRT06765.1 KRT06766.1 OUUW01000003 SPP78516.1 CH963925 EDW78269.2 KRF98844.1 JXUM01056431 KQ561910 KXJ77198.1 CH916371 EDV91912.1 KQ434931 KZC11845.1 KQ414615 KOC68757.1 GDKW01000749 JAI55846.1 KQ760382 OAD60700.1 CH940651 EDW65857.1 KRF82444.1 CP012528 ALC49112.1 GGFM01002934 MBW23685.1 KQ435714 KOX79211.1 UFQS01000201 UFQT01000201 SSX01253.1 SSX21633.1 GEZM01096133 JAV55135.1 KQ971345 EFA05107.2 AJWK01011058
Proteomes
UP000005204
UP000283053
UP000053268
UP000218220
UP000007151
UP000075886
+ More
UP000076408 UP000075881 UP000075901 UP000075884 UP000075920 UP000075903 UP000075885 UP000075902 UP000076407 UP000075900 UP000007062 UP000075882 UP000075840 UP000075883 UP000030765 UP000075880 UP000235965 UP000002358 UP000027135 UP000215335 UP000008820 UP000000673 UP000279307 UP000053097 UP000092443 UP000078200 UP000007801 UP000002282 UP000092444 UP000008711 UP000092445 UP000069272 UP000000803 UP000092460 UP000192221 UP000000311 UP000001819 UP000242457 UP000008237 UP000268350 UP000007798 UP000005203 UP000069940 UP000249989 UP000001070 UP000076502 UP000053825 UP000008792 UP000092553 UP000053105 UP000007266 UP000092461
UP000076408 UP000075881 UP000075901 UP000075884 UP000075920 UP000075903 UP000075885 UP000075902 UP000076407 UP000075900 UP000007062 UP000075882 UP000075840 UP000075883 UP000030765 UP000075880 UP000235965 UP000002358 UP000027135 UP000215335 UP000008820 UP000000673 UP000279307 UP000053097 UP000092443 UP000078200 UP000007801 UP000002282 UP000092444 UP000008711 UP000092445 UP000069272 UP000000803 UP000092460 UP000192221 UP000000311 UP000001819 UP000242457 UP000008237 UP000268350 UP000007798 UP000005203 UP000069940 UP000249989 UP000001070 UP000076502 UP000053825 UP000008792 UP000092553 UP000053105 UP000007266 UP000092461
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J880
A0A3S2M7Y4
A0A194PWR1
A0A2A4JPV3
A0A2H1WST7
A0A212EJ44
+ More
A0A182QD14 A0A182XZM3 A0A182K4Z9 A0A182T7L3 A0A182MZC1 A0A1Y9IUX2 A0A182UXA3 A0A1S4GX10 A0A182P0D4 A0A182TST7 A0A1Y9IVL3 A0A182X1I7 A0A182RU97 Q7Q3B4 A0A182L2I9 A0A182IEG8 A0A1Y9IXE8 A0A182MER4 A0A084VI29 A0A240PLB3 A0A182J972 A0A2J7RDW6 A0A0C9S1N5 A0A1Q3G4C0 K7IVN1 A0A1Q3G4E4 A0A0C9RMP5 A0A1Q3G4D3 A0A1Q3G4A4 A0A1Q3G4C3 A0A067REZ8 A0A232ENX7 W8BGE7 Q16Q77 A0A1S4FSY8 W8C2N2 W5J4G0 A0A3L8E1J0 A0A2M4CG58 A0A026WJ96 A0A2M4CGE5 A0A2M4A6X3 A0A1B6E866 A0A1A9YGV4 A0A1A9UV25 A0A1B6JFG3 A0A1Q3G491 B3MQK6 B4PYY9 A0A0R1EAQ4 A0A0P8Y4K7 A0A1B0FM21 A0A0P8YKQ7 B3NWE8 A0A1B0AJ08 A0A1I8JSH2 Q9W5Y0 A0A1B0AT16 A0A0Q5T447 A0A1W4UQT4 A0A182F1R2 A0A1Q3G498 A0A1W4UR29 Q8IS64 E1ZWD4 A0A2M4BEE5 A0A2M4BED4 A0A0R3P4T4 A0A2A3EQ90 Q29GV1 A0A023F3J7 E2B3X1 A0A0R3P5C3 A0A0R3P0H4 A0A3B0JYR1 B4N1N0 A0A0Q9X3K1 A0A087ZS76 A0A182GDM3 B4JMG9 A0A154PJ26 A0A0L7RCP1 A0A0P4VL55 A0A310SFY3 B4M2F3 A0A0Q9WCR8 A0A0M4F9B9 A0A2M3Z5A3 A0A0N0BJI3 A0A336LY78 A0A1Y1K5Z5 D2A597 A0A1B0CGE7
A0A182QD14 A0A182XZM3 A0A182K4Z9 A0A182T7L3 A0A182MZC1 A0A1Y9IUX2 A0A182UXA3 A0A1S4GX10 A0A182P0D4 A0A182TST7 A0A1Y9IVL3 A0A182X1I7 A0A182RU97 Q7Q3B4 A0A182L2I9 A0A182IEG8 A0A1Y9IXE8 A0A182MER4 A0A084VI29 A0A240PLB3 A0A182J972 A0A2J7RDW6 A0A0C9S1N5 A0A1Q3G4C0 K7IVN1 A0A1Q3G4E4 A0A0C9RMP5 A0A1Q3G4D3 A0A1Q3G4A4 A0A1Q3G4C3 A0A067REZ8 A0A232ENX7 W8BGE7 Q16Q77 A0A1S4FSY8 W8C2N2 W5J4G0 A0A3L8E1J0 A0A2M4CG58 A0A026WJ96 A0A2M4CGE5 A0A2M4A6X3 A0A1B6E866 A0A1A9YGV4 A0A1A9UV25 A0A1B6JFG3 A0A1Q3G491 B3MQK6 B4PYY9 A0A0R1EAQ4 A0A0P8Y4K7 A0A1B0FM21 A0A0P8YKQ7 B3NWE8 A0A1B0AJ08 A0A1I8JSH2 Q9W5Y0 A0A1B0AT16 A0A0Q5T447 A0A1W4UQT4 A0A182F1R2 A0A1Q3G498 A0A1W4UR29 Q8IS64 E1ZWD4 A0A2M4BEE5 A0A2M4BED4 A0A0R3P4T4 A0A2A3EQ90 Q29GV1 A0A023F3J7 E2B3X1 A0A0R3P5C3 A0A0R3P0H4 A0A3B0JYR1 B4N1N0 A0A0Q9X3K1 A0A087ZS76 A0A182GDM3 B4JMG9 A0A154PJ26 A0A0L7RCP1 A0A0P4VL55 A0A310SFY3 B4M2F3 A0A0Q9WCR8 A0A0M4F9B9 A0A2M3Z5A3 A0A0N0BJI3 A0A336LY78 A0A1Y1K5Z5 D2A597 A0A1B0CGE7
PDB
3DWB
E-value=0,
Score=1653
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell membrane
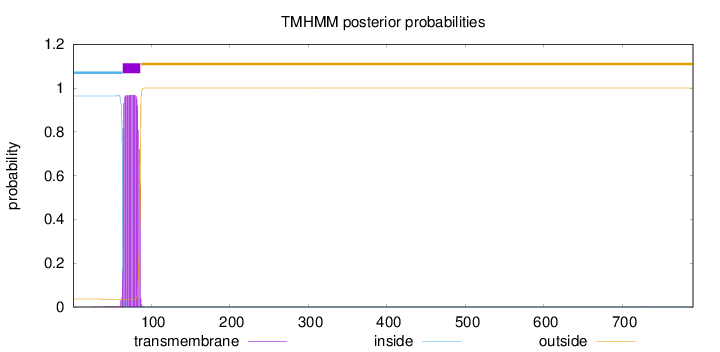
Length:
790
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.53308
Exp number, first 60 AAs:
0.06227
Total prob of N-in:
0.96363
inside
1 - 63
TMhelix
64 - 86
outside
87 - 790
Population Genetic Test Statistics
Pi
185.23701
Theta
157.846155
Tajima's D
0.321105
CLR
0.852207
CSRT
0.460626968651567
Interpretation
Uncertain
