Gene
KWMTBOMO15961 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005717
Annotation
PREDICTED:_dihydropyrimidinase_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.83
Sequence
CDS
ATGACCGTTGACGCCACAGCGACATCTGTGTCCGGTGTTAATATCAAAAGCTCTCAAAACCGGCTTCTGATCAAGAATGGCAGGATCGTTAACGACGATGGTATCGAGGAGGCTGACGTCTACATTGAAGACGGCATCATCAAGCAAGTTGGCCGCAACCTCATCATTCCTGGAGGCACACGGACCATCGATGCCGCTGGCAAACTGGTCATGCCCGGGGGCATCGACCCCCACACTCACTTCGAGCTGGAAATGATGGGCGCTAAAACTGCCGATGACTTCTACAAGGGCACCCGGGCTGCGGTCGCTGGAGGGACAACAACCATAATTGACTTCGTTCTACCTGAAAAGGGGCAGTCGCTTCTAGAGGCCTATGCGAATTGGCGGGAAAAAGCCGATAATAAGGTGTGCTGCGACTACGGTCTCCACGTAGGAGTGACCTGGTGGTCGCCGGCCGTGCAGAAGGAGATGAAACAGCTGGTCTCCGAGCAGTACGGCGTGAACTCCTTCAAGATGTTCATGGCGTACCGGAACACCTGGATGCTCGACGACTACGACCTGTACTGCGCCATCGAGACCTGTGCTGAACTCAAAGCCCTGCCCCAAGTCCACGCAGAGAACGGCCACATCATAGCTCGCAACACCGAGAAGCTCCTCGCGGCCGGAGTGACCAGGCCCGAGGGTCATCAGCTCTCCAGAGACGAGGAAGTCGAAAGCGAGGCCGTGAACAGGGCATGCGTCATCGCCAATCAGGCAAACAGCCCCCTGTATATCGTGCACATGATGTCGAAGAGCGCAGTCCGAGCTCTCCAACTGGCCCGCAACAAACAGAAACAGCCTGTATACGGGGAAACTCTGGCCGCCACTGTCGGTACCGATGGTACTCACTACCGGAACTCCTGTTTCCGTCACGCGGCCGCTCACGTGCTCTCTCCTCCACTGAGACCTGACCCGTCCACTCCTCAAGCCATGGTGGAAGCGCTGGCCGATGACCACTTACAGCTGATCGGCAGCGACAACTGCACGTTCCACGAGAAAGACAAGGAAATCGGCAAAGGGAACTTCTCCAAGATTCCGAATGGCGTTAATGGAGTCGAAGACCGCATGACCATATTGTGGCAGAAGGGAGTCAACACTGGAATAATGGACCCGTGCCGGTTCGTGGCGGTGACCAGCTCCAGCGCCGCTAAGATCTTCAACCTGTGGCCGCAGAAGGGGCGCGTGGCGGTCAACAGCGACGCCGACATCGTCGTCTGGGATCCGCGCCTCGAGAAGACCATAACCGCCGCCAGCCACAAGCAGGCGGTCGACTTCAACATCTTCGAGGGTCAGCACGTGGTCGGCGGGCCGCAGTACGTCATCGTGAACGGCAGGGTGTGCCTGGACGACGGCGAACTCAGAGTCGTTGAAGGTCTAGGCAAGTTCCTGAAGACCCCAATAAACAGTCCGTACGTTTACGGGGAGCAGAATCTATCGCACGGCGACCACGACCTGCTCGAGCCGCTAAGCCGGGATTCCCCCGATAGGGTCAACGGTTCCTCCGTGCCAACCGACCTGGCCCTCGTGAACAAGCATCTCGAAGGTCTGAGCGTGACGTCATCCGGCTGTAGCACCCCGACCGGACGGAAAATGAGGGACCCCATGCAGAGGGACTTGCAAAACTCCACCTTCTCTATCAGCAAAGAGACTGAAGGACTCGACACCAAGACTTCGGTTCGCGTCCGCAACCCGCCGGGCGGCAAGTCCTCCGGGCTCTGGTAG
Protein
MTVDATATSVSGVNIKSSQNRLLIKNGRIVNDDGIEEADVYIEDGIIKQVGRNLIIPGGTRTIDAAGKLVMPGGIDPHTHFELEMMGAKTADDFYKGTRAAVAGGTTTIIDFVLPEKGQSLLEAYANWREKADNKVCCDYGLHVGVTWWSPAVQKEMKQLVSEQYGVNSFKMFMAYRNTWMLDDYDLYCAIETCAELKALPQVHAENGHIIARNTEKLLAAGVTRPEGHQLSRDEEVESEAVNRACVIANQANSPLYIVHMMSKSAVRALQLARNKQKQPVYGETLAATVGTDGTHYRNSCFRHAAAHVLSPPLRPDPSTPQAMVEALADDHLQLIGSDNCTFHEKDKEIGKGNFSKIPNGVNGVEDRMTILWQKGVNTGIMDPCRFVAVTSSSAAKIFNLWPQKGRVAVNSDADIVVWDPRLEKTITAASHKQAVDFNIFEGQHVVGGPQYVIVNGRVCLDDGELRVVEGLGKFLKTPINSPYVYGEQNLSHGDHDLLEPLSRDSPDRVNGSSVPTDLALVNKHLEGLSVTSSGCSTPTGRKMRDPMQRDLQNSTFSISKETEGLDTKTSVRVRNPPGGKSSGLW
Summary
Uniprot
A0A3S2P022
A0A2H1W4V5
A0A0N0PE68
A0A194Q221
A0A2A4KAT6
A0A212FBK2
+ More
A0A1S4FHC5 A0A023EVC9 A0A1Y1JWL0 A0A1Q3FZ81 U5ERH4 A0A1L8DY05 A0A1L8DXV1 A0A0P6GVF2 A0A0P6DZ65 A0A0P5E6K1 A0A0P5T0U9 A0A0P5VLS1 A0A026VXA1 E9HEG7 A0A0P6AA33 A0A0P5VU65 A0A3F2YSP3 A0A2M4CIC6 A0A0P5TAY5 A0A158N946 A0A0P6BI46 W5JG30 A0A0P5K9R4 A0A151IQ59 A0A0P5T3M7 A0A0P5ALC3 A0A0P5YQZ7 A0A0P5A115 A0A1B6DMF4 A0A0K8W795 A0A034WEF9 A0A0K8VS20 A0A1B6LJK0 A0A0N8ABD8 A0A0P5ZK81 A0A034WIJ3 A0A0P5D6G8 A0A0P5FGC6 A0A1I8NWP1 A0A0P5UYD8 K7J5X3 A0A0N8CGY8 A0A0N8CHE4 A0A0P4ZJ43 A0A0P5BLY7 J9JRJ8 A0A0P5K0T4 A0A0P5V4Z7 A0A336KUI5 A0A0C9RYA2 A0A0N8CYM0 A0A0T6AUZ7 D6W7R7 B4KU98 A0A2J7RMY9 A0A2H8U0N4 A0A0P5NJJ5 A0A1Y1K493 A0A1Q3G4S4 J9EA44 A0A1B6IG16 A0A1B6F4W7 Q171I1 B4LKF4 A0A1S4FH89 A0A0P5UQE7 A0A336M9S2 A0A1B6DF52 K1PGY5 V4AUM4 A0A0P5FYR3 V9IGU4 A0A3R7M0S1 B4NFK4 A0A0P5KNS2 A0A0P4XMH6 B4J537 A0A1I8MJV5 A0A1I8MBF6 A0A0P5T385 T1PGN2 A0A0P4WCD9 A0A1Q3G4L9 A0A182XVL4 A0A0P4WL42 A0A0M3QV77 A0A210PIS8 B3N1T5 A0A1W4UY79 A0A0P5KM06 A0A1S3D1S6 Q8IPQ2 A0A1Y1K2S8
A0A1S4FHC5 A0A023EVC9 A0A1Y1JWL0 A0A1Q3FZ81 U5ERH4 A0A1L8DY05 A0A1L8DXV1 A0A0P6GVF2 A0A0P6DZ65 A0A0P5E6K1 A0A0P5T0U9 A0A0P5VLS1 A0A026VXA1 E9HEG7 A0A0P6AA33 A0A0P5VU65 A0A3F2YSP3 A0A2M4CIC6 A0A0P5TAY5 A0A158N946 A0A0P6BI46 W5JG30 A0A0P5K9R4 A0A151IQ59 A0A0P5T3M7 A0A0P5ALC3 A0A0P5YQZ7 A0A0P5A115 A0A1B6DMF4 A0A0K8W795 A0A034WEF9 A0A0K8VS20 A0A1B6LJK0 A0A0N8ABD8 A0A0P5ZK81 A0A034WIJ3 A0A0P5D6G8 A0A0P5FGC6 A0A1I8NWP1 A0A0P5UYD8 K7J5X3 A0A0N8CGY8 A0A0N8CHE4 A0A0P4ZJ43 A0A0P5BLY7 J9JRJ8 A0A0P5K0T4 A0A0P5V4Z7 A0A336KUI5 A0A0C9RYA2 A0A0N8CYM0 A0A0T6AUZ7 D6W7R7 B4KU98 A0A2J7RMY9 A0A2H8U0N4 A0A0P5NJJ5 A0A1Y1K493 A0A1Q3G4S4 J9EA44 A0A1B6IG16 A0A1B6F4W7 Q171I1 B4LKF4 A0A1S4FH89 A0A0P5UQE7 A0A336M9S2 A0A1B6DF52 K1PGY5 V4AUM4 A0A0P5FYR3 V9IGU4 A0A3R7M0S1 B4NFK4 A0A0P5KNS2 A0A0P4XMH6 B4J537 A0A1I8MJV5 A0A1I8MBF6 A0A0P5T385 T1PGN2 A0A0P4WCD9 A0A1Q3G4L9 A0A182XVL4 A0A0P4WL42 A0A0M3QV77 A0A210PIS8 B3N1T5 A0A1W4UY79 A0A0P5KM06 A0A1S3D1S6 Q8IPQ2 A0A1Y1K2S8
Pubmed
EMBL
RSAL01000012
RVE53514.1
ODYU01006314
SOQ48053.1
KQ459933
KPJ19275.1
+ More
KQ459590 KPI97455.1 NWSH01000003 PCG81024.1 AGBW02009300 OWR51124.1 GAPW01000657 JAC12941.1 GEZM01098876 JAV53692.1 GFDL01002198 JAV32847.1 GANO01002806 JAB57065.1 GFDF01002795 JAV11289.1 GFDF01002794 JAV11290.1 GDIQ01028206 JAN66531.1 GDIQ01070105 JAN24632.1 GDIP01147389 JAJ76013.1 GDIP01137186 JAL66528.1 GDIP01099585 JAM04130.1 KK107638 EZA48392.1 GL732629 EFX69844.1 GDIP01045266 LRGB01003271 JAM58449.1 KZS03445.1 GDIP01109796 JAL93918.1 APCN01002114 GGFL01000723 MBW64901.1 GDIP01132742 JAL70972.1 ADTU01009076 ADTU01009077 ADTU01009078 ADTU01009079 GDIP01014374 JAM89341.1 ADMH02001318 ETN63026.1 GDIQ01187406 JAK64319.1 KQ976785 KYN08291.1 GDIP01131496 JAL72218.1 GDIP01211077 JAJ12325.1 GDIP01056008 JAM47707.1 GDIP01209869 JAJ13533.1 GEDC01010460 JAS26838.1 GDHF01027016 GDHF01018802 GDHF01017488 GDHF01005390 GDHF01004659 JAI25298.1 JAI33512.1 JAI34826.1 JAI46924.1 JAI47655.1 GAKP01004991 GAKP01004989 JAC53961.1 GDHF01010623 GDHF01006962 JAI41691.1 JAI45352.1 GEBQ01016149 JAT23828.1 GDIP01164033 JAJ59369.1 GDIP01056009 JAM47706.1 GAKP01004992 JAC53960.1 GDIP01164786 JAJ58616.1 GDIQ01254930 JAJ96794.1 GDIP01109797 JAL93917.1 GDIP01132743 JAL70971.1 GDIP01131495 JAL72219.1 GDIP01212077 JAJ11325.1 GDIP01186725 JAJ36677.1 ABLF02034884 ABLF02034890 ABLF02034903 GDIQ01191456 JAK60269.1 GDIP01104947 JAL98767.1 UFQS01000940 UFQT01000940 SSX07882.1 SSX28116.1 GBYB01012926 JAG82693.1 GDIP01086087 JAM17628.1 LJIG01022826 KRT78519.1 KQ971307 EFA11336.2 CH933808 EDW09694.1 NEVH01002547 PNF42198.1 GFXV01007945 MBW19750.1 GDIQ01145376 JAL06350.1 GEZM01098879 JAV53687.1 GFDL01000276 JAV34769.1 CH477454 EJY57696.1 GECU01021853 JAS85853.1 GECZ01024542 JAS45227.1 EAT40651.1 CH940648 EDW60675.1 GDIP01109800 JAL93914.1 UFQS01000755 UFQT01000755 SSX06649.1 SSX26995.1 GEDC01013028 JAS24270.1 JH817585 EKC20858.1 KB201305 ESO97476.1 GDIQ01253839 JAJ97885.1 JR047070 AEY60318.1 QCYY01002657 ROT68566.1 CH964251 EDW83071.2 GDIQ01188854 JAK62871.1 GDIP01240482 JAI82919.1 CH916367 EDW00663.1 GDIP01133993 JAL69721.1 KA647300 AFP61929.1 GDRN01081560 JAI61994.1 GFDL01000295 JAV34750.1 GDRN01081563 JAI61993.1 CP012524 ALC41903.1 NEDP02076628 OWF36385.1 CH902661 EDV34054.2 GDIQ01187407 JAK64318.1 AE014297 AF465756 GU452674 GU452675 AAN13245.2 AAO33382.1 ADD13532.1 GEZM01098880 JAV53686.1
KQ459590 KPI97455.1 NWSH01000003 PCG81024.1 AGBW02009300 OWR51124.1 GAPW01000657 JAC12941.1 GEZM01098876 JAV53692.1 GFDL01002198 JAV32847.1 GANO01002806 JAB57065.1 GFDF01002795 JAV11289.1 GFDF01002794 JAV11290.1 GDIQ01028206 JAN66531.1 GDIQ01070105 JAN24632.1 GDIP01147389 JAJ76013.1 GDIP01137186 JAL66528.1 GDIP01099585 JAM04130.1 KK107638 EZA48392.1 GL732629 EFX69844.1 GDIP01045266 LRGB01003271 JAM58449.1 KZS03445.1 GDIP01109796 JAL93918.1 APCN01002114 GGFL01000723 MBW64901.1 GDIP01132742 JAL70972.1 ADTU01009076 ADTU01009077 ADTU01009078 ADTU01009079 GDIP01014374 JAM89341.1 ADMH02001318 ETN63026.1 GDIQ01187406 JAK64319.1 KQ976785 KYN08291.1 GDIP01131496 JAL72218.1 GDIP01211077 JAJ12325.1 GDIP01056008 JAM47707.1 GDIP01209869 JAJ13533.1 GEDC01010460 JAS26838.1 GDHF01027016 GDHF01018802 GDHF01017488 GDHF01005390 GDHF01004659 JAI25298.1 JAI33512.1 JAI34826.1 JAI46924.1 JAI47655.1 GAKP01004991 GAKP01004989 JAC53961.1 GDHF01010623 GDHF01006962 JAI41691.1 JAI45352.1 GEBQ01016149 JAT23828.1 GDIP01164033 JAJ59369.1 GDIP01056009 JAM47706.1 GAKP01004992 JAC53960.1 GDIP01164786 JAJ58616.1 GDIQ01254930 JAJ96794.1 GDIP01109797 JAL93917.1 GDIP01132743 JAL70971.1 GDIP01131495 JAL72219.1 GDIP01212077 JAJ11325.1 GDIP01186725 JAJ36677.1 ABLF02034884 ABLF02034890 ABLF02034903 GDIQ01191456 JAK60269.1 GDIP01104947 JAL98767.1 UFQS01000940 UFQT01000940 SSX07882.1 SSX28116.1 GBYB01012926 JAG82693.1 GDIP01086087 JAM17628.1 LJIG01022826 KRT78519.1 KQ971307 EFA11336.2 CH933808 EDW09694.1 NEVH01002547 PNF42198.1 GFXV01007945 MBW19750.1 GDIQ01145376 JAL06350.1 GEZM01098879 JAV53687.1 GFDL01000276 JAV34769.1 CH477454 EJY57696.1 GECU01021853 JAS85853.1 GECZ01024542 JAS45227.1 EAT40651.1 CH940648 EDW60675.1 GDIP01109800 JAL93914.1 UFQS01000755 UFQT01000755 SSX06649.1 SSX26995.1 GEDC01013028 JAS24270.1 JH817585 EKC20858.1 KB201305 ESO97476.1 GDIQ01253839 JAJ97885.1 JR047070 AEY60318.1 QCYY01002657 ROT68566.1 CH964251 EDW83071.2 GDIQ01188854 JAK62871.1 GDIP01240482 JAI82919.1 CH916367 EDW00663.1 GDIP01133993 JAL69721.1 KA647300 AFP61929.1 GDRN01081560 JAI61994.1 GFDL01000295 JAV34750.1 GDRN01081563 JAI61993.1 CP012524 ALC41903.1 NEDP02076628 OWF36385.1 CH902661 EDV34054.2 GDIQ01187407 JAK64318.1 AE014297 AF465756 GU452674 GU452675 AAN13245.2 AAO33382.1 ADD13532.1 GEZM01098880 JAV53686.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000218220
UP000007151
UP000053097
+ More
UP000000305 UP000076858 UP000075840 UP000005205 UP000000673 UP000078542 UP000095300 UP000002358 UP000007819 UP000007266 UP000009192 UP000235965 UP000008820 UP000008792 UP000005408 UP000030746 UP000283509 UP000007798 UP000001070 UP000095301 UP000076408 UP000092553 UP000242188 UP000007801 UP000192221 UP000079169 UP000000803
UP000000305 UP000076858 UP000075840 UP000005205 UP000000673 UP000078542 UP000095300 UP000002358 UP000007819 UP000007266 UP000009192 UP000235965 UP000008820 UP000008792 UP000005408 UP000030746 UP000283509 UP000007798 UP000001070 UP000095301 UP000076408 UP000092553 UP000242188 UP000007801 UP000192221 UP000079169 UP000000803
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A3S2P022
A0A2H1W4V5
A0A0N0PE68
A0A194Q221
A0A2A4KAT6
A0A212FBK2
+ More
A0A1S4FHC5 A0A023EVC9 A0A1Y1JWL0 A0A1Q3FZ81 U5ERH4 A0A1L8DY05 A0A1L8DXV1 A0A0P6GVF2 A0A0P6DZ65 A0A0P5E6K1 A0A0P5T0U9 A0A0P5VLS1 A0A026VXA1 E9HEG7 A0A0P6AA33 A0A0P5VU65 A0A3F2YSP3 A0A2M4CIC6 A0A0P5TAY5 A0A158N946 A0A0P6BI46 W5JG30 A0A0P5K9R4 A0A151IQ59 A0A0P5T3M7 A0A0P5ALC3 A0A0P5YQZ7 A0A0P5A115 A0A1B6DMF4 A0A0K8W795 A0A034WEF9 A0A0K8VS20 A0A1B6LJK0 A0A0N8ABD8 A0A0P5ZK81 A0A034WIJ3 A0A0P5D6G8 A0A0P5FGC6 A0A1I8NWP1 A0A0P5UYD8 K7J5X3 A0A0N8CGY8 A0A0N8CHE4 A0A0P4ZJ43 A0A0P5BLY7 J9JRJ8 A0A0P5K0T4 A0A0P5V4Z7 A0A336KUI5 A0A0C9RYA2 A0A0N8CYM0 A0A0T6AUZ7 D6W7R7 B4KU98 A0A2J7RMY9 A0A2H8U0N4 A0A0P5NJJ5 A0A1Y1K493 A0A1Q3G4S4 J9EA44 A0A1B6IG16 A0A1B6F4W7 Q171I1 B4LKF4 A0A1S4FH89 A0A0P5UQE7 A0A336M9S2 A0A1B6DF52 K1PGY5 V4AUM4 A0A0P5FYR3 V9IGU4 A0A3R7M0S1 B4NFK4 A0A0P5KNS2 A0A0P4XMH6 B4J537 A0A1I8MJV5 A0A1I8MBF6 A0A0P5T385 T1PGN2 A0A0P4WCD9 A0A1Q3G4L9 A0A182XVL4 A0A0P4WL42 A0A0M3QV77 A0A210PIS8 B3N1T5 A0A1W4UY79 A0A0P5KM06 A0A1S3D1S6 Q8IPQ2 A0A1Y1K2S8
A0A1S4FHC5 A0A023EVC9 A0A1Y1JWL0 A0A1Q3FZ81 U5ERH4 A0A1L8DY05 A0A1L8DXV1 A0A0P6GVF2 A0A0P6DZ65 A0A0P5E6K1 A0A0P5T0U9 A0A0P5VLS1 A0A026VXA1 E9HEG7 A0A0P6AA33 A0A0P5VU65 A0A3F2YSP3 A0A2M4CIC6 A0A0P5TAY5 A0A158N946 A0A0P6BI46 W5JG30 A0A0P5K9R4 A0A151IQ59 A0A0P5T3M7 A0A0P5ALC3 A0A0P5YQZ7 A0A0P5A115 A0A1B6DMF4 A0A0K8W795 A0A034WEF9 A0A0K8VS20 A0A1B6LJK0 A0A0N8ABD8 A0A0P5ZK81 A0A034WIJ3 A0A0P5D6G8 A0A0P5FGC6 A0A1I8NWP1 A0A0P5UYD8 K7J5X3 A0A0N8CGY8 A0A0N8CHE4 A0A0P4ZJ43 A0A0P5BLY7 J9JRJ8 A0A0P5K0T4 A0A0P5V4Z7 A0A336KUI5 A0A0C9RYA2 A0A0N8CYM0 A0A0T6AUZ7 D6W7R7 B4KU98 A0A2J7RMY9 A0A2H8U0N4 A0A0P5NJJ5 A0A1Y1K493 A0A1Q3G4S4 J9EA44 A0A1B6IG16 A0A1B6F4W7 Q171I1 B4LKF4 A0A1S4FH89 A0A0P5UQE7 A0A336M9S2 A0A1B6DF52 K1PGY5 V4AUM4 A0A0P5FYR3 V9IGU4 A0A3R7M0S1 B4NFK4 A0A0P5KNS2 A0A0P4XMH6 B4J537 A0A1I8MJV5 A0A1I8MBF6 A0A0P5T385 T1PGN2 A0A0P4WCD9 A0A1Q3G4L9 A0A182XVL4 A0A0P4WL42 A0A0M3QV77 A0A210PIS8 B3N1T5 A0A1W4UY79 A0A0P5KM06 A0A1S3D1S6 Q8IPQ2 A0A1Y1K2S8
PDB
2VR2
E-value=1.07786e-151,
Score=1378
Ontologies
PATHWAY
00240
Pyrimidine metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00770 Pantothenate and CoA biosynthesis - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00770 Pantothenate and CoA biosynthesis - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
Topology
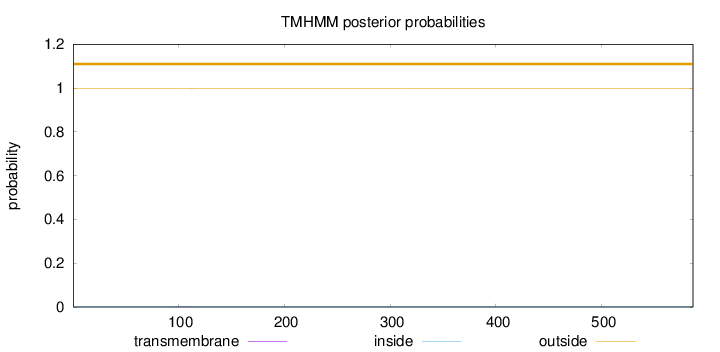
Length:
586
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0343900000000002
Exp number, first 60 AAs:
0.00029
Total prob of N-in:
0.00363
outside
1 - 586
Population Genetic Test Statistics
Pi
280.270725
Theta
165.105749
Tajima's D
2.632036
CLR
0.250794
CSRT
0.95705214739263
Interpretation
Uncertain
