Pre Gene Modal
BGIBMGA013427
Annotation
PREDICTED:_uncharacterized_protein_LOC106123133_isoform_X5_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.747 Nuclear Reliability : 1.074
Sequence
CDS
ATGACGACGGAGAACGCGACGCCTGGCCAGGCGGACGCCAGCACGGCGTTCCTACGCGCTGCCCGTGCCGGAAACATCGAGAAGATCATCAGTCTTCTCGAGCAAGGCGTTGATATCAACGTCAGCAATGCGAATGGCCTAAATGCCATCCATCTCGCTTCCAAAGACGGCCATGTCGAAGTAGTGAGGGTCCTGCTGGAGCGAGGAGCAGCAATCGACGCGGCCACCAAGAAAGGGAACACAGCACTTCATATTGCTTCTCTGGCTGGTCAAGAGGCAGTCGTTAAACTACTCGTCCAAAATGGAGCTCAGGTCAACATCCAATCCCAGAATGGTTTCACACCTTTATACATGGCAGCCCAAGAGAACCACGACGGTGTGGTCAAGTTTCTTCTTGGTAACGGTGCGAACCAGAGCCTTGCCACTGAGGACGGATTCACGCCTCTAGCGGTGGCGATGCAACAAGGACACGAGAAAGTAGTGGCAGTTCTTTTAGAGGCAGACACGCGCGGTCGAGTGCGACTACCAGCCCTTCACATTGCAGCCAAGAAAGACGACGTGAAGGCTGCTAACTTACTCTTAGAGAACGAGCACAATCCAGACGTGACCTCTAAGTCTGGATTCACTCCGCTACACATCGCGGCTCACTACGGCAACGAAGCAGTCGCAAGACTATTGCTGGCCAAAGGCGCTGACGTCAACTGTGCTGCGAAACACAACATATGTCCGTTACACGTCGCTGCCAAATGGGGCAAGGAGAATATGGTAGCTTTGTTATGCGATAATGGGGCGAATGTAGAGGCCCGCACTAGAGATGGGCTGACCCCTCTACACTGTGCCGCCAGATCTGGCCACGAGAGAGTCGTTGAAGCTTTACTTGATAGGGGTGCTCCTATTACCAGTAAGAGCAAGAACGGTCTAGCCCCGTTGCACATGTCGGCGCAAGGCGAACACGCCGAGGCGGCGCGCGTGCTCTTGTCGCGACGCGCCCCCGTCGATGACGTCACCGTGGACTACCTCACCGCGCTTCATGTAGCCGCTCACTGCGGTCACGCTAAAGTCGCGAAACTGCTTTTAGATAGAAATGCCGACGCGAACGCTAGAGCCTTGAATGGCTTCACACCTTTGCACATCGCCTGCAAGAAAAATAGGATCAAGGTCGTTGAGTTACTCCTCAAATACGGTGCAAGTATCCAGGCAACAACAGAATCAGGTCTTACGCCCCTCCACGTGGCGGCTTTTATGGGCTGCATGAACATCGTGATTTACTTACTCCAGCACGAGGCGAATCCTGATGTTCCCACGGTCAGGGGAGAGACACCGCTACATTTGGCAGCAAGAGCTAACCAGACTGACATTATCCGTATTCTTCTACGTAACGGCGCGGCCGTCGAAGCCAAGGCTCGTGAGAGGCAAACCCCTCTCCACATCGCGTCTCGGCTCGGTAACGTCGACATCGCAGTACTGCTGTTACAGCACGGCGCTGACGTCAGAGCCATTACTGCTGACCATTACAATCCACTGCACATCGCTGCCAAACAGCACAATCACGATGTTGCAGCAGCTTTAATTGAACACAACGCACCCCTAACTGCGACCACAAAGAAAGGGTTCACCGCTCTTCATCTGGCTGCGAAGTATGGGAACCTGAAGGTTGCGAATCTTTTACTCGCTCACGGTGCGGCTCCAGACCAAGCTGGTAAAAATGGCATGACTCCTCTACACATCGCTGCACAGTACGACCAGCAAGCCGTCGCCACTACGCTCCTTGAGAAAGGAGCTGATGCTAAGGCTGTAGCCAAGAACGGGCATACACCTCTTCATATCGCGGCTCGTAAGAATCAAATGGAAACTGCAGCCACGCTTCTCGAGTACGGAGCGTTGACCAACGCTGAATCAAAGGCTGGATTTACTCCTCTGCATTTGGCTGCCCAGCAGGGTCACACCGAAATGTGCGCTCTGCTTCTCGAACATAGCGCTGAACCAGAGCATCAGTCTAAAAACGGGCTGGCAGCATTGCATTTGGCTGCTCAAGAAGATAAGGTCCCAGTAGCACAACTGCTAGTGAAGCATGGCGCTGAGGTGGACATCTGCACTAAGGGCGGCTACACGCCTCTACACATTGCCAGTCACTATGGCCAACCGAACATGGTGCGCTATTTACTCGAAAATGGTGCTTCTGTTAAAGCTCAAACCGCACATGGTTACACAGCCCTACATCACGCCGCTCAGCAGGGCCACATTAATATTGTGAATATTCTACTTGAACACAAAGCAGATGCAAACGCGACGACTACGAGCGGACAAACTCCTTTGGATATAGCGTCCAAGCTGGGTTACGTAACAGTGATGGAGACTCTTAAGGAAATATCAGAGCCGTCGTCGGCACCAGCCTCGCAGGAAAAGTACAAGGTGGTCGCCCCGGAGACCATGCTTGAGACCTTCATGTCTGACTCAGAGGAAGAAGGTGGTGAAGACACCATTCTAAACGACCAGCCATACAGATATCTGACCGCCGATGATATGAAGTCTTTGGGAGATGATTCTCTGCCAATCGACGTGACTAAGGATGAACGAACAGAATCCGCCATGAGCCATAAGAATATCATGGAAATCTCTCAAGGGTCTATGAACGGGATTCCGTACCAACCTCAGCAAGAAGTGGTGGTGAAGAGCATTAGTCGTTACAGTACTGCACAGGCTCCTGAGGCGTATTGCTACAACTGA
Protein
MTTENATPGQADASTAFLRAARAGNIEKIISLLEQGVDINVSNANGLNAIHLASKDGHVEVVRVLLERGAAIDAATKKGNTALHIASLAGQEAVVKLLVQNGAQVNIQSQNGFTPLYMAAQENHDGVVKFLLGNGANQSLATEDGFTPLAVAMQQGHEKVVAVLLEADTRGRVRLPALHIAAKKDDVKAANLLLENEHNPDVTSKSGFTPLHIAAHYGNEAVARLLLAKGADVNCAAKHNICPLHVAAKWGKENMVALLCDNGANVEARTRDGLTPLHCAARSGHERVVEALLDRGAPITSKSKNGLAPLHMSAQGEHAEAARVLLSRRAPVDDVTVDYLTALHVAAHCGHAKVAKLLLDRNADANARALNGFTPLHIACKKNRIKVVELLLKYGASIQATTESGLTPLHVAAFMGCMNIVIYLLQHEANPDVPTVRGETPLHLAARANQTDIIRILLRNGAAVEAKARERQTPLHIASRLGNVDIAVLLLQHGADVRAITADHYNPLHIAAKQHNHDVAAALIEHNAPLTATTKKGFTALHLAAKYGNLKVANLLLAHGAAPDQAGKNGMTPLHIAAQYDQQAVATTLLEKGADAKAVAKNGHTPLHIAARKNQMETAATLLEYGALTNAESKAGFTPLHLAAQQGHTEMCALLLEHSAEPEHQSKNGLAALHLAAQEDKVPVAQLLVKHGAEVDICTKGGYTPLHIASHYGQPNMVRYLLENGASVKAQTAHGYTALHHAAQQGHINIVNILLEHKADANATTTSGQTPLDIASKLGYVTVMETLKEISEPSSAPASQEKYKVVAPETMLETFMSDSEEEGGEDTILNDQPYRYLTADDMKSLGDDSLPIDVTKDERTESAMSHKNIMEISQGSMNGIPYQPQQEVVVKSISRYSTAQAPEAYCYN
Summary
Uniprot
H9JV64
A0A3S2NQA0
A0A2A4KAM8
A0A2H1W4F3
A0A194PWR8
A0A1S4G7S4
+ More
Q16PP6 A0A0R1DZP0 A0A2J7QN88 A0A0R1DWE6 A0A1W4WTJ8 A0A0R1DW73 Q7KU92 A8JNM6 M9MRX1 A0A0P8XVJ4 B3M617 A0A0N8P124 M9MRR4 A0A0R1E1F7 A0A0R1DWE4 A0A0R1DW69 B4PCA6 A0A0P8YIB7 A0A182FT56 A0A1W4VME5 A0A1W4VN49 A8JNM4 M9MS19 X2JGD3 A0A0R1DW90 A0A0R1E149 A0A0R1DXZ9 A0A0P8XVD6 Q7KU95 A0A2J7QN90 A0A0Q5UDJ7 F5HIS2 A0A182ND69 A0A0N8P123 A0A0P8XVE8 X2JC49 M9PEN3 A0A182Y4S7 F5HIS1 A0A0J9RQT6 A0A0J9UGH1 A0A0J9RR56 A0A1W4VAD9 A0A1W4VNS8 A0A182K4G2 A0A1B6G621 A0A182QUQ5 A0A0R1DWU2 A0A0J9UGF0 A0A1I8P656 A0A0Q9XBT7 A0A195CXE2 A0A1W4VN44 A0A1W4VAQ1 A0A0J9RR51 A8JNM7 A0A0J9RSA7 A0A0J9RQT0 A0A0Q9XCL9 A0A1B0G162 A0A0R3P372 A0A151WHJ1 A0A1I8MJB4 A0A182R2C7 A0A0P8YBE2 A0A0P9C4S2 A0A0P9ALJ9 A0A182VQ03 M9MSK6 A0A0R1DW55 A0A0K8U7Y0 A0A182X3C2 Q9NCP8 A0A0P8XVK6 Q7QKD3 A8JNM5 A0A1W4WHD6 A0A1B6HTP3 A0A0J9RQN0 A0A0J9RR40 A0A1B6HGE1 A0A336MKZ5 A0A1I8P653 A0A182PLK8 A0A0A1XHM9 A0A0J9RQP1 A0A1I8P647 A0A0K8U6U5 A0A1I8MJB1 A0A0J9UGG5 A0A1A9XCG6 A0A1W4VAQ7 A0A139WKB6 B4IX91
Q16PP6 A0A0R1DZP0 A0A2J7QN88 A0A0R1DWE6 A0A1W4WTJ8 A0A0R1DW73 Q7KU92 A8JNM6 M9MRX1 A0A0P8XVJ4 B3M617 A0A0N8P124 M9MRR4 A0A0R1E1F7 A0A0R1DWE4 A0A0R1DW69 B4PCA6 A0A0P8YIB7 A0A182FT56 A0A1W4VME5 A0A1W4VN49 A8JNM4 M9MS19 X2JGD3 A0A0R1DW90 A0A0R1E149 A0A0R1DXZ9 A0A0P8XVD6 Q7KU95 A0A2J7QN90 A0A0Q5UDJ7 F5HIS2 A0A182ND69 A0A0N8P123 A0A0P8XVE8 X2JC49 M9PEN3 A0A182Y4S7 F5HIS1 A0A0J9RQT6 A0A0J9UGH1 A0A0J9RR56 A0A1W4VAD9 A0A1W4VNS8 A0A182K4G2 A0A1B6G621 A0A182QUQ5 A0A0R1DWU2 A0A0J9UGF0 A0A1I8P656 A0A0Q9XBT7 A0A195CXE2 A0A1W4VN44 A0A1W4VAQ1 A0A0J9RR51 A8JNM7 A0A0J9RSA7 A0A0J9RQT0 A0A0Q9XCL9 A0A1B0G162 A0A0R3P372 A0A151WHJ1 A0A1I8MJB4 A0A182R2C7 A0A0P8YBE2 A0A0P9C4S2 A0A0P9ALJ9 A0A182VQ03 M9MSK6 A0A0R1DW55 A0A0K8U7Y0 A0A182X3C2 Q9NCP8 A0A0P8XVK6 Q7QKD3 A8JNM5 A0A1W4WHD6 A0A1B6HTP3 A0A0J9RQN0 A0A0J9RR40 A0A1B6HGE1 A0A336MKZ5 A0A1I8P653 A0A182PLK8 A0A0A1XHM9 A0A0J9RQP1 A0A1I8P647 A0A0K8U6U5 A0A1I8MJB1 A0A0J9UGG5 A0A1A9XCG6 A0A1W4VAQ7 A0A139WKB6 B4IX91
Pubmed
EMBL
BABH01026485
BABH01026486
BABH01026487
RSAL01000012
RVE53517.1
NWSH01000003
+ More
PCG81036.1 ODYU01006276 SOQ47970.1 KQ459590 KPI97463.1 CH477775 EAT36335.1 CM000159 KRK01379.1 NEVH01013198 PNF30054.1 KRK01369.1 KRK01376.1 AE014296 AAF50525.4 ABW08487.1 ADV37501.1 CH902618 KPU78721.1 EDV40733.2 KPU78732.1 ADV37505.1 KRK01370.1 KRK01373.1 KRK01371.1 EDW93791.2 KPU78726.1 ABW08486.1 ADV37506.1 AHN57995.1 KRK01372.1 KRK01377.1 KRK01374.1 KPU78725.1 AAN12046.2 PNF30051.1 CH954178 KQS43297.1 AAAB01008799 EGK96183.1 KPU78724.1 KPU78734.1 AHN57996.1 AGB94230.1 EGK96182.1 CM002912 KMY98183.1 KMY98195.1 KMY98192.1 GECZ01011883 JAS57886.1 AXCN02001215 KRK01375.1 KMY98175.1 CH933809 KRG06006.1 KQ977220 KYN04819.1 KMY98187.1 ABW08485.1 KMY98194.1 KMY98178.1 KRG06005.1 CCAG010020781 CH379069 KRT07608.1 KQ983114 KYQ47306.1 KPU78729.1 KPU78727.1 KPU78722.1 ADV37504.1 KRK01368.1 GDHF01033775 GDHF01029616 GDHF01012932 JAI18539.1 JAI22698.1 JAI39382.1 AF190635 BT133155 AAF73309.1 ABW08483.1 AEZ02848.1 KPU78730.1 EAA03765.4 ABW08488.1 GECU01029664 JAS78042.1 KMY98181.1 KMY98177.1 GECU01033946 JAS73760.1 UFQS01000711 UFQT01000711 SSX06344.1 SSX26698.1 GBXI01003801 JAD10491.1 KMY98191.1 GDHF01030234 GDHF01000529 JAI22080.1 JAI51785.1 KMY98190.1 KQ971332 KYB28281.1 CH916366 EDV97423.1
PCG81036.1 ODYU01006276 SOQ47970.1 KQ459590 KPI97463.1 CH477775 EAT36335.1 CM000159 KRK01379.1 NEVH01013198 PNF30054.1 KRK01369.1 KRK01376.1 AE014296 AAF50525.4 ABW08487.1 ADV37501.1 CH902618 KPU78721.1 EDV40733.2 KPU78732.1 ADV37505.1 KRK01370.1 KRK01373.1 KRK01371.1 EDW93791.2 KPU78726.1 ABW08486.1 ADV37506.1 AHN57995.1 KRK01372.1 KRK01377.1 KRK01374.1 KPU78725.1 AAN12046.2 PNF30051.1 CH954178 KQS43297.1 AAAB01008799 EGK96183.1 KPU78724.1 KPU78734.1 AHN57996.1 AGB94230.1 EGK96182.1 CM002912 KMY98183.1 KMY98195.1 KMY98192.1 GECZ01011883 JAS57886.1 AXCN02001215 KRK01375.1 KMY98175.1 CH933809 KRG06006.1 KQ977220 KYN04819.1 KMY98187.1 ABW08485.1 KMY98194.1 KMY98178.1 KRG06005.1 CCAG010020781 CH379069 KRT07608.1 KQ983114 KYQ47306.1 KPU78729.1 KPU78727.1 KPU78722.1 ADV37504.1 KRK01368.1 GDHF01033775 GDHF01029616 GDHF01012932 JAI18539.1 JAI22698.1 JAI39382.1 AF190635 BT133155 AAF73309.1 ABW08483.1 AEZ02848.1 KPU78730.1 EAA03765.4 ABW08488.1 GECU01029664 JAS78042.1 KMY98181.1 KMY98177.1 GECU01033946 JAS73760.1 UFQS01000711 UFQT01000711 SSX06344.1 SSX26698.1 GBXI01003801 JAD10491.1 KMY98191.1 GDHF01030234 GDHF01000529 JAI22080.1 JAI51785.1 KMY98190.1 KQ971332 KYB28281.1 CH916366 EDV97423.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000008820
UP000002282
+ More
UP000235965 UP000192223 UP000000803 UP000007801 UP000069272 UP000192221 UP000008711 UP000007062 UP000075884 UP000076408 UP000075881 UP000075886 UP000095300 UP000009192 UP000078542 UP000092444 UP000001819 UP000075809 UP000095301 UP000075900 UP000075920 UP000076407 UP000075885 UP000092443 UP000007266 UP000001070
UP000235965 UP000192223 UP000000803 UP000007801 UP000069272 UP000192221 UP000008711 UP000007062 UP000075884 UP000076408 UP000075881 UP000075886 UP000095300 UP000009192 UP000078542 UP000092444 UP000001819 UP000075809 UP000095301 UP000075900 UP000075920 UP000076407 UP000075885 UP000092443 UP000007266 UP000001070
Pfam
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JV64
A0A3S2NQA0
A0A2A4KAM8
A0A2H1W4F3
A0A194PWR8
A0A1S4G7S4
+ More
Q16PP6 A0A0R1DZP0 A0A2J7QN88 A0A0R1DWE6 A0A1W4WTJ8 A0A0R1DW73 Q7KU92 A8JNM6 M9MRX1 A0A0P8XVJ4 B3M617 A0A0N8P124 M9MRR4 A0A0R1E1F7 A0A0R1DWE4 A0A0R1DW69 B4PCA6 A0A0P8YIB7 A0A182FT56 A0A1W4VME5 A0A1W4VN49 A8JNM4 M9MS19 X2JGD3 A0A0R1DW90 A0A0R1E149 A0A0R1DXZ9 A0A0P8XVD6 Q7KU95 A0A2J7QN90 A0A0Q5UDJ7 F5HIS2 A0A182ND69 A0A0N8P123 A0A0P8XVE8 X2JC49 M9PEN3 A0A182Y4S7 F5HIS1 A0A0J9RQT6 A0A0J9UGH1 A0A0J9RR56 A0A1W4VAD9 A0A1W4VNS8 A0A182K4G2 A0A1B6G621 A0A182QUQ5 A0A0R1DWU2 A0A0J9UGF0 A0A1I8P656 A0A0Q9XBT7 A0A195CXE2 A0A1W4VN44 A0A1W4VAQ1 A0A0J9RR51 A8JNM7 A0A0J9RSA7 A0A0J9RQT0 A0A0Q9XCL9 A0A1B0G162 A0A0R3P372 A0A151WHJ1 A0A1I8MJB4 A0A182R2C7 A0A0P8YBE2 A0A0P9C4S2 A0A0P9ALJ9 A0A182VQ03 M9MSK6 A0A0R1DW55 A0A0K8U7Y0 A0A182X3C2 Q9NCP8 A0A0P8XVK6 Q7QKD3 A8JNM5 A0A1W4WHD6 A0A1B6HTP3 A0A0J9RQN0 A0A0J9RR40 A0A1B6HGE1 A0A336MKZ5 A0A1I8P653 A0A182PLK8 A0A0A1XHM9 A0A0J9RQP1 A0A1I8P647 A0A0K8U6U5 A0A1I8MJB1 A0A0J9UGG5 A0A1A9XCG6 A0A1W4VAQ7 A0A139WKB6 B4IX91
Q16PP6 A0A0R1DZP0 A0A2J7QN88 A0A0R1DWE6 A0A1W4WTJ8 A0A0R1DW73 Q7KU92 A8JNM6 M9MRX1 A0A0P8XVJ4 B3M617 A0A0N8P124 M9MRR4 A0A0R1E1F7 A0A0R1DWE4 A0A0R1DW69 B4PCA6 A0A0P8YIB7 A0A182FT56 A0A1W4VME5 A0A1W4VN49 A8JNM4 M9MS19 X2JGD3 A0A0R1DW90 A0A0R1E149 A0A0R1DXZ9 A0A0P8XVD6 Q7KU95 A0A2J7QN90 A0A0Q5UDJ7 F5HIS2 A0A182ND69 A0A0N8P123 A0A0P8XVE8 X2JC49 M9PEN3 A0A182Y4S7 F5HIS1 A0A0J9RQT6 A0A0J9UGH1 A0A0J9RR56 A0A1W4VAD9 A0A1W4VNS8 A0A182K4G2 A0A1B6G621 A0A182QUQ5 A0A0R1DWU2 A0A0J9UGF0 A0A1I8P656 A0A0Q9XBT7 A0A195CXE2 A0A1W4VN44 A0A1W4VAQ1 A0A0J9RR51 A8JNM7 A0A0J9RSA7 A0A0J9RQT0 A0A0Q9XCL9 A0A1B0G162 A0A0R3P372 A0A151WHJ1 A0A1I8MJB4 A0A182R2C7 A0A0P8YBE2 A0A0P9C4S2 A0A0P9ALJ9 A0A182VQ03 M9MSK6 A0A0R1DW55 A0A0K8U7Y0 A0A182X3C2 Q9NCP8 A0A0P8XVK6 Q7QKD3 A8JNM5 A0A1W4WHD6 A0A1B6HTP3 A0A0J9RQN0 A0A0J9RR40 A0A1B6HGE1 A0A336MKZ5 A0A1I8P653 A0A182PLK8 A0A0A1XHM9 A0A0J9RQP1 A0A1I8P647 A0A0K8U6U5 A0A1I8MJB1 A0A0J9UGG5 A0A1A9XCG6 A0A1W4VAQ7 A0A139WKB6 B4IX91
PDB
4RLV
E-value=0,
Score=2458
Ontologies
GO
GO:0007165
GO:0000226
GO:0005886
GO:0007605
GO:0031594
GO:0043195
GO:0045887
GO:0036062
GO:0007528
GO:0042734
GO:1900074
GO:0008092
GO:0007614
GO:0048675
GO:0070050
GO:0006869
GO:0005576
GO:0042157
GO:0008289
GO:0016021
GO:0005515
GO:0005525
GO:0007264
GO:0006811
GO:0005220
GO:0005783
GO:0006816
GO:0000287
GO:0006281
GO:0006259
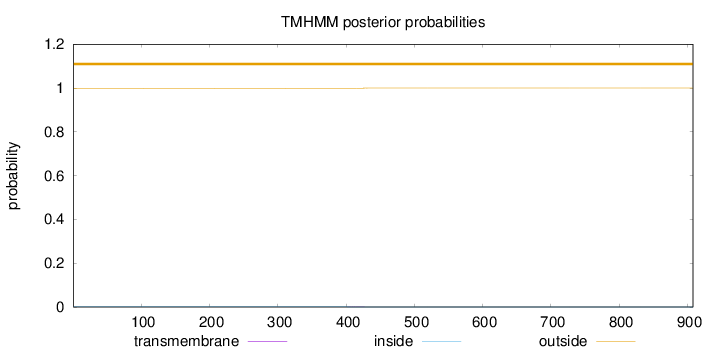
Topology
Length:
908
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01839
Exp number, first 60 AAs:
0.0001
Total prob of N-in:
0.00077
outside
1 - 908
Population Genetic Test Statistics
Pi
288.902645
Theta
173.506502
Tajima's D
2.063803
CLR
0.365661
CSRT
0.891805409729513
Interpretation
Uncertain
