Gene
KWMTBOMO15948
Pre Gene Modal
BGIBMGA013424
Annotation
hypothetical_protein_KGM_03254_[Danaus_plexippus]
Full name
Rho GTPase-activating protein 190
Alternative Name
Rho GTPase-activating protein of 190 kDa
Location in the cell
Extracellular Reliability : 1.114 Nuclear Reliability : 1.33
Sequence
CDS
ATGTCGTTGTATGTTTGTAGAGTCGCCGACAACTCGAAACTGAACTCGATGGACAGCAAGAACCTGGCGATATGCTGGTGGCCGACCCTCCTCCCGGTGCAGTTCTCGGACATGGGCAGGTTCGAGATGACGAGGCCCTACCTCGAGGACATCGTCCAGACAATGATCGATCAGCACCCGTTTCTGTTTCGAGGGCAGGAGGCGTTTGTCATGGTGTGA
Protein
MSLYVCRVADNSKLNSMDSKNLAICWWPTLLPVQFSDMGRFEMTRPYLEDIVQTMIDQHPFLFRGQEAFVMV
Summary
Description
GTPase-activating protein (GAP) for RhoA/Rho1 that plays an essential role in the stability of dorsal branches of mushroom body (MB) neurons. The MB neurons are the center for olfactory learning and memory. Acts by converting RhoA/Rho1 to an inactive GDP-bound state, leading to repress the RhoA/Rho1-Drok-MRLC signaling pathway thereby maintaining axon branch stability.
Keywords
Alternative splicing
Coiled coil
Complete proteome
Developmental protein
GTPase activation
Phosphoprotein
Reference proteome
Repeat
Feature
chain Rho GTPase-activating protein 190
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A0N0PCX5
A0A194PVW5
A0A3S2TRZ6
A0A0L7LR77
H9JV61
A0A2A4JSL4
+ More
A0A1Y1KQ67 A0A1W4XDQ3 A0A1Y1KQ68 A0A1Y1KTM3 A0A182J7I3 A0A1Y1KVX8 A0A1Y1KSX7 A0A182U865 A0A1S3DII1 A0A139WPB0 A0A139WPM0 D6W848 A0A139WPL6 A0A139WPB3 A0A1B6GH64 A0A1B6IEV2 A0A139WPT7 A0A1B6H8T2 A0A1B6K9C5 A0A1B6LPU4 A0A1B6D9I5 A0A1B6DTL1 A0A1B6C2R3 A0A182HMS3 A0A182PWH5 A0A182K6L6 A0A182N8J0 Q7Q4U2 A0A1B6CUS5 A0A182QWI7 A0A182XYA6 A0A182R261 A0A1J1HUZ3 A0A1J1HT50 W5JMC7 A0A1B0FY42 A0A2M4B8P1 A0A182FJM9 A0A2M4B923 A0A2M4A9K4 A0A182VVF3 A0A182M261 E0VLS6 A0A087ZTU0 A0A2A3EBP9 A0A0L7RBB4 A0A1I8PE60 A0A1I8PE53 A0A1I8PE48 E2C5J7 A0A1I8PE55 A0A1I8PEE1 A0A310SAT8 A0A1Y1KQ63 A0A232EWD7 A0A0T6B326 A0A0Q9WDF1 A0A0Q9WQX0 A0A0Q9WPF4 A0A0Q9WPQ1 B4MAZ2 A0A0Q9WE01 A0A0Q9WNN5 A0A0M3QZ95 A0A0R3NXW7 A0A026WFE0 A0A3B0J7L7 A0A3B0JBC8 A0A0R3P2Z7 A0A0R3P3J5 A0A0R3P485 A0A0R3P3N5 A0A3B0J6L2 B4H9W9 A0A3B0JWS2 A0A0R1EI73 B5DKM2 A0A0Q5TAE3 A0A0Q5T3Z0 M9PJQ3 A0A0R3NY69 A0A0R3NXX8 A0A1B0A2U6 A0A1B0FZZ7 A0A0C5KMR0 B4Q2I7 Q9VX32 A0A0Q5T3R5 A0A0R1EBJ3 A0A1A9URJ8 A0A0Q9XE92 A0A0Q9XSK8 A0A0Q5T6H5 M9PHY4
A0A1Y1KQ67 A0A1W4XDQ3 A0A1Y1KQ68 A0A1Y1KTM3 A0A182J7I3 A0A1Y1KVX8 A0A1Y1KSX7 A0A182U865 A0A1S3DII1 A0A139WPB0 A0A139WPM0 D6W848 A0A139WPL6 A0A139WPB3 A0A1B6GH64 A0A1B6IEV2 A0A139WPT7 A0A1B6H8T2 A0A1B6K9C5 A0A1B6LPU4 A0A1B6D9I5 A0A1B6DTL1 A0A1B6C2R3 A0A182HMS3 A0A182PWH5 A0A182K6L6 A0A182N8J0 Q7Q4U2 A0A1B6CUS5 A0A182QWI7 A0A182XYA6 A0A182R261 A0A1J1HUZ3 A0A1J1HT50 W5JMC7 A0A1B0FY42 A0A2M4B8P1 A0A182FJM9 A0A2M4B923 A0A2M4A9K4 A0A182VVF3 A0A182M261 E0VLS6 A0A087ZTU0 A0A2A3EBP9 A0A0L7RBB4 A0A1I8PE60 A0A1I8PE53 A0A1I8PE48 E2C5J7 A0A1I8PE55 A0A1I8PEE1 A0A310SAT8 A0A1Y1KQ63 A0A232EWD7 A0A0T6B326 A0A0Q9WDF1 A0A0Q9WQX0 A0A0Q9WPF4 A0A0Q9WPQ1 B4MAZ2 A0A0Q9WE01 A0A0Q9WNN5 A0A0M3QZ95 A0A0R3NXW7 A0A026WFE0 A0A3B0J7L7 A0A3B0JBC8 A0A0R3P2Z7 A0A0R3P3J5 A0A0R3P485 A0A0R3P3N5 A0A3B0J6L2 B4H9W9 A0A3B0JWS2 A0A0R1EI73 B5DKM2 A0A0Q5TAE3 A0A0Q5T3Z0 M9PJQ3 A0A0R3NY69 A0A0R3NXX8 A0A1B0A2U6 A0A1B0FZZ7 A0A0C5KMR0 B4Q2I7 Q9VX32 A0A0Q5T3R5 A0A0R1EBJ3 A0A1A9URJ8 A0A0Q9XE92 A0A0Q9XSK8 A0A0Q5T6H5 M9PHY4
Pubmed
26354079
26227816
19121390
28004739
18362917
19820115
+ More
12364791 14747013 17210077 25244985 20920257 23761445 20566863 20798317 28648823 17994087 18057021 15632085 24508170 23185243 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11672527 12537569 17227793 18327897
12364791 14747013 17210077 25244985 20920257 23761445 20566863 20798317 28648823 17994087 18057021 15632085 24508170 23185243 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11672527 12537569 17227793 18327897
EMBL
KQ460448
KPJ14685.1
KQ459590
KPI97467.1
RSAL01000012
RVE53521.1
+ More
JTDY01000278 KOB77945.1 BABH01026464 NWSH01000653 PCG75001.1 GEZM01076910 JAV63559.1 GEZM01076914 JAV63552.1 GEZM01076909 JAV63560.1 GEZM01076912 JAV63556.1 GEZM01076915 JAV63551.1 KQ971307 KYB29839.1 KYB29836.1 EFA10998.2 KYB29837.1 KYB29838.1 GECZ01007981 JAS61788.1 GECU01022284 JAS85422.1 KYB29835.1 GECU01036667 JAS71039.1 GEBQ01031930 JAT08047.1 GEBQ01014443 JAT25534.1 GEDC01014951 JAS22347.1 GEDC01008319 JAS28979.1 GEDC01029497 JAS07801.1 APCN01004886 AAAB01008963 EAA12077.5 GEDC01020009 JAS17289.1 AXCN02002019 AXCN02002020 CVRI01000020 CRK91180.1 CRK91181.1 ADMH02000691 ETN65301.1 AJVK01005168 GGFJ01000229 MBW49370.1 GGFJ01000392 MBW49533.1 GGFK01004163 MBW37484.1 AXCM01004922 DS235281 EEB14332.1 KZ288291 PBC29203.1 KQ414617 KOC68212.1 GL452770 EFN76810.1 KQ778189 OAD52014.1 GEZM01076913 GEZM01076911 JAV63553.1 NNAY01001878 OXU22660.1 LJIG01016036 KRT81808.1 CH940655 KRF82668.1 KRF82672.1 KRF82665.1 KRF82669.1 KRF82670.1 KRF82667.1 EDW66401.2 KRF82666.1 KRF82671.1 CP012528 ALC49033.1 CH379063 KRT05922.1 KK107274 EZA53739.1 OUUW01000003 SPP77805.1 SPP77803.1 KRT05925.1 KRT05920.1 KRT05927.1 KRT05926.1 KRT05923.1 SPP77804.1 CH479232 EDW36626.1 SPP77806.1 CM000162 KRK06866.1 EDY72072.1 CH954180 KQS29998.1 KQS30001.1 KQS29997.1 AE014298 AGB95501.1 KRT05924.1 KRT05921.1 CCAG010006727 BT150459 AJP62081.1 EDX02628.1 KRK06860.1 AF387518 AY121666 KQS29994.1 KQS30002.1 KRK06863.1 KRK06864.1 CH933811 KRG06892.1 KRG06899.1 KQS30000.1 AGB95499.1
JTDY01000278 KOB77945.1 BABH01026464 NWSH01000653 PCG75001.1 GEZM01076910 JAV63559.1 GEZM01076914 JAV63552.1 GEZM01076909 JAV63560.1 GEZM01076912 JAV63556.1 GEZM01076915 JAV63551.1 KQ971307 KYB29839.1 KYB29836.1 EFA10998.2 KYB29837.1 KYB29838.1 GECZ01007981 JAS61788.1 GECU01022284 JAS85422.1 KYB29835.1 GECU01036667 JAS71039.1 GEBQ01031930 JAT08047.1 GEBQ01014443 JAT25534.1 GEDC01014951 JAS22347.1 GEDC01008319 JAS28979.1 GEDC01029497 JAS07801.1 APCN01004886 AAAB01008963 EAA12077.5 GEDC01020009 JAS17289.1 AXCN02002019 AXCN02002020 CVRI01000020 CRK91180.1 CRK91181.1 ADMH02000691 ETN65301.1 AJVK01005168 GGFJ01000229 MBW49370.1 GGFJ01000392 MBW49533.1 GGFK01004163 MBW37484.1 AXCM01004922 DS235281 EEB14332.1 KZ288291 PBC29203.1 KQ414617 KOC68212.1 GL452770 EFN76810.1 KQ778189 OAD52014.1 GEZM01076913 GEZM01076911 JAV63553.1 NNAY01001878 OXU22660.1 LJIG01016036 KRT81808.1 CH940655 KRF82668.1 KRF82672.1 KRF82665.1 KRF82669.1 KRF82670.1 KRF82667.1 EDW66401.2 KRF82666.1 KRF82671.1 CP012528 ALC49033.1 CH379063 KRT05922.1 KK107274 EZA53739.1 OUUW01000003 SPP77805.1 SPP77803.1 KRT05925.1 KRT05920.1 KRT05927.1 KRT05926.1 KRT05923.1 SPP77804.1 CH479232 EDW36626.1 SPP77806.1 CM000162 KRK06866.1 EDY72072.1 CH954180 KQS29998.1 KQS30001.1 KQS29997.1 AE014298 AGB95501.1 KRT05924.1 KRT05921.1 CCAG010006727 BT150459 AJP62081.1 EDX02628.1 KRK06860.1 AF387518 AY121666 KQS29994.1 KQS30002.1 KRK06863.1 KRK06864.1 CH933811 KRG06892.1 KRG06899.1 KQS30000.1 AGB95499.1
Proteomes
UP000053240
UP000053268
UP000283053
UP000037510
UP000005204
UP000218220
+ More
UP000192223 UP000075880 UP000075902 UP000079169 UP000007266 UP000075840 UP000075885 UP000075881 UP000075884 UP000007062 UP000075886 UP000076408 UP000075900 UP000183832 UP000000673 UP000092462 UP000069272 UP000075920 UP000075883 UP000009046 UP000005203 UP000242457 UP000053825 UP000095300 UP000008237 UP000215335 UP000008792 UP000092553 UP000001819 UP000053097 UP000268350 UP000008744 UP000002282 UP000008711 UP000000803 UP000092445 UP000092444 UP000078200 UP000009192
UP000192223 UP000075880 UP000075902 UP000079169 UP000007266 UP000075840 UP000075885 UP000075881 UP000075884 UP000007062 UP000075886 UP000076408 UP000075900 UP000183832 UP000000673 UP000092462 UP000069272 UP000075920 UP000075883 UP000009046 UP000005203 UP000242457 UP000053825 UP000095300 UP000008237 UP000215335 UP000008792 UP000092553 UP000001819 UP000053097 UP000268350 UP000008744 UP000002282 UP000008711 UP000000803 UP000092445 UP000092444 UP000078200 UP000009192
Interpro
Gene 3D
ProteinModelPortal
A0A0N0PCX5
A0A194PVW5
A0A3S2TRZ6
A0A0L7LR77
H9JV61
A0A2A4JSL4
+ More
A0A1Y1KQ67 A0A1W4XDQ3 A0A1Y1KQ68 A0A1Y1KTM3 A0A182J7I3 A0A1Y1KVX8 A0A1Y1KSX7 A0A182U865 A0A1S3DII1 A0A139WPB0 A0A139WPM0 D6W848 A0A139WPL6 A0A139WPB3 A0A1B6GH64 A0A1B6IEV2 A0A139WPT7 A0A1B6H8T2 A0A1B6K9C5 A0A1B6LPU4 A0A1B6D9I5 A0A1B6DTL1 A0A1B6C2R3 A0A182HMS3 A0A182PWH5 A0A182K6L6 A0A182N8J0 Q7Q4U2 A0A1B6CUS5 A0A182QWI7 A0A182XYA6 A0A182R261 A0A1J1HUZ3 A0A1J1HT50 W5JMC7 A0A1B0FY42 A0A2M4B8P1 A0A182FJM9 A0A2M4B923 A0A2M4A9K4 A0A182VVF3 A0A182M261 E0VLS6 A0A087ZTU0 A0A2A3EBP9 A0A0L7RBB4 A0A1I8PE60 A0A1I8PE53 A0A1I8PE48 E2C5J7 A0A1I8PE55 A0A1I8PEE1 A0A310SAT8 A0A1Y1KQ63 A0A232EWD7 A0A0T6B326 A0A0Q9WDF1 A0A0Q9WQX0 A0A0Q9WPF4 A0A0Q9WPQ1 B4MAZ2 A0A0Q9WE01 A0A0Q9WNN5 A0A0M3QZ95 A0A0R3NXW7 A0A026WFE0 A0A3B0J7L7 A0A3B0JBC8 A0A0R3P2Z7 A0A0R3P3J5 A0A0R3P485 A0A0R3P3N5 A0A3B0J6L2 B4H9W9 A0A3B0JWS2 A0A0R1EI73 B5DKM2 A0A0Q5TAE3 A0A0Q5T3Z0 M9PJQ3 A0A0R3NY69 A0A0R3NXX8 A0A1B0A2U6 A0A1B0FZZ7 A0A0C5KMR0 B4Q2I7 Q9VX32 A0A0Q5T3R5 A0A0R1EBJ3 A0A1A9URJ8 A0A0Q9XE92 A0A0Q9XSK8 A0A0Q5T6H5 M9PHY4
A0A1Y1KQ67 A0A1W4XDQ3 A0A1Y1KQ68 A0A1Y1KTM3 A0A182J7I3 A0A1Y1KVX8 A0A1Y1KSX7 A0A182U865 A0A1S3DII1 A0A139WPB0 A0A139WPM0 D6W848 A0A139WPL6 A0A139WPB3 A0A1B6GH64 A0A1B6IEV2 A0A139WPT7 A0A1B6H8T2 A0A1B6K9C5 A0A1B6LPU4 A0A1B6D9I5 A0A1B6DTL1 A0A1B6C2R3 A0A182HMS3 A0A182PWH5 A0A182K6L6 A0A182N8J0 Q7Q4U2 A0A1B6CUS5 A0A182QWI7 A0A182XYA6 A0A182R261 A0A1J1HUZ3 A0A1J1HT50 W5JMC7 A0A1B0FY42 A0A2M4B8P1 A0A182FJM9 A0A2M4B923 A0A2M4A9K4 A0A182VVF3 A0A182M261 E0VLS6 A0A087ZTU0 A0A2A3EBP9 A0A0L7RBB4 A0A1I8PE60 A0A1I8PE53 A0A1I8PE48 E2C5J7 A0A1I8PE55 A0A1I8PEE1 A0A310SAT8 A0A1Y1KQ63 A0A232EWD7 A0A0T6B326 A0A0Q9WDF1 A0A0Q9WQX0 A0A0Q9WPF4 A0A0Q9WPQ1 B4MAZ2 A0A0Q9WE01 A0A0Q9WNN5 A0A0M3QZ95 A0A0R3NXW7 A0A026WFE0 A0A3B0J7L7 A0A3B0JBC8 A0A0R3P2Z7 A0A0R3P3J5 A0A0R3P485 A0A0R3P3N5 A0A3B0J6L2 B4H9W9 A0A3B0JWS2 A0A0R1EI73 B5DKM2 A0A0Q5TAE3 A0A0Q5T3Z0 M9PJQ3 A0A0R3NY69 A0A0R3NXX8 A0A1B0A2U6 A0A1B0FZZ7 A0A0C5KMR0 B4Q2I7 Q9VX32 A0A0Q5T3R5 A0A0R1EBJ3 A0A1A9URJ8 A0A0Q9XE92 A0A0Q9XSK8 A0A0Q5T6H5 M9PHY4
PDB
5IRC
E-value=1.05336e-08,
Score=136
Ontologies
GO
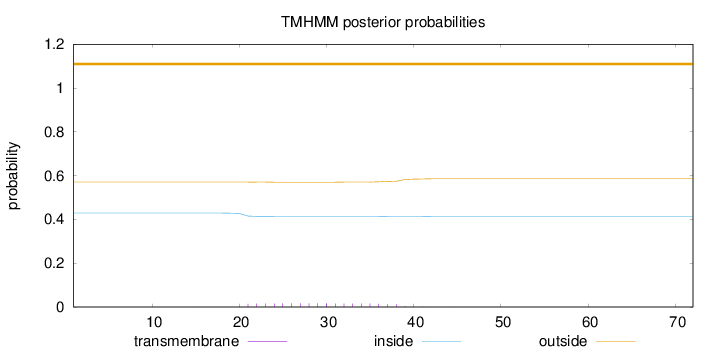
Topology
Length:
72
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.29046
Exp number, first 60 AAs:
0.29046
Total prob of N-in:
0.42934
outside
1 - 72
Population Genetic Test Statistics
Pi
291.687347
Theta
203.601477
Tajima's D
1.339455
CLR
0.043805
CSRT
0.747862606869657
Interpretation
Uncertain
