Pre Gene Modal
BGIBMGA013423
Annotation
PREDICTED:_rho_GTPase-activating_protein_190_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.301
Sequence
CDS
ATGGCAAAAAAATCTGATAATACTGGCGGGAAGCTGGTTACCATATCTGTAGTTGGTTTGTCTGGTACGGAAAAGGAGAAAGGGCAGCTAGGCGTGGGCAAGTCGTGCCTCTGTAACAGGTTTGTTCGAACCCACGCCGATGATTACAACGTCGATCACATTTGCGTTCTGAGTCAGTCGGATTTCAGCGGGCGCGTTGTGAACAACGATCATTTCCTGTACTGGGGCAACGTGCAGAAGGAGAACGACGATCAGGAGTACAGGTTCGAGATAGTCGAGCAGACCGAGTTTGTGGACGATGCGTGCTTTCAGCCGTTCAAAGTTGGCAAAACTGAACCGTACGTGAAGCGATGCGTCGCCACGAAACTCCAGTCGGCCGAGAAATACATGTACGTGTGCAAGAACCAGCTCGGCATAGAGAAGGAGTACGAGCAGAAACTGATGCCGGAAGGGAAGCTCACCGTTGACGGGTTCATGTGTGTCTATGACGTCAGCCTGGTGCCGGGGAGGACTTGGGAGAAACAGAACGAAATTATAGCGGCCATATTGCAGAATATAATCAAGCTCAAGAAGCCGGTTGTACTGGTGACTACGAAAAACGATGAGGCTTGCGACCAGGGAGTCCGGGAGGCCGAACGCTTAGTTCAAAGGAAAGAGTTCAAAGGTTCCATACCGATAGTGGAGACGTCCAGCCACGACAACGTCAATGTGGACCAAGCGTTCTTCCTGCTGGCCCAGATGATTGATAAGTCGAAAGCGAGGATCAAGGTGGCGAACTACGCGGAGGCTCTGCGCATAAGGCGGGAAACTTTGGACTTCGTGACGGAAGCGTTCACTCAGCTCATACGGATACACGTCGAGGATCATAAAGAATTATGGTCTTCTGTGAGCAAGAGGCTTTGTCATCACGCGGAGTGGATTAAGTTTGTTCAACAGTTTGGGAATGATGGTACTCAAGTCGTGTTCCGTCGGCACGTGCGCCGCCTCAAGGAGGAGCGCTCTGCGAAGAAGCTGCGGAACCAACTCGCCAAGCTGCCTCAAGTGTTGGCGAGACTCCAGCTGCCCACCGAAGAACTACAAGAGAACGACTGGCCGGTGGTGGTGCGGCAGCTGCGCCTGCACCGCGACTTCTCGGTGTACTTCAGCGAGGGCCGCGGCGCCGACTCGTGCTCCGAGACCGGCTCCGAGCTGGACAGCCCCCTCGCCATCGACACCACGCTGCGGGTCGGCGAGAGGACTCGCTATCAGACACTCGGTCAGTCGCAGAAGATACCCTACGAGATATTGGAGACGAACGAGGCAGCTGCGGTGTTCAAGTCCTTCCTTCACGAAGCCCAGGAGGAGCAGAGGAATTATGAATGGTGTCAGCAATTCAAACGTCTCCTGGAGGAGACCGGGTACGTGACCCCTGGGAAGCAGCTGTCGGAGGTGCGCGTGCTGCTGATGGGCAGGGAGTGCTACGAGGCCCTCTCCGAAGAACAACAGCAGAGGGTCTACGACCAGCACCAGAGGCAGATACAGCGTCGGGCGAAACACAACTTTCAGGAGTTGCTACTGGAGCACGCGGACCTGTTCTACCATTTCAAGAGCATATCGCCGACCGGGACCATAACGCAAGAAGACATCAAGGAAATCACCGATGTGCTGCAAGATGACTTTAGGTACAAAATGCTGGATCGGATGGAGCAAGACAGGAAGCTGATGCTGTTCCAGCACCTCGGGTTCGTGCACTGCCCGATGCGGGAGCACTGCCCCGCCGGACCCAACTGTCTGGACGCCACCCTGCCCGTCATACTCAACACCAGAGTCGGGTCGCTGACGTCGGCAGGCGAGTCCCAGTGCCTGGCCGGGCCGCCCGCCGCGCCCTGGGCCCTCACCACCGACACCAACCACATCAACGTCATCATCCTCGGGATGGAAGGTGTGGCTGGCGAATTCGGCAAGCGCCTGATGGCCGGCTGCGACTCCGAGCGGCGCATCAACGTGCACGGACACAGCTGGAGGCTCGAGCAGAGGATCAGGACCGATGACTTCTCTGAAACGACCTCCATAGACGATTTTGCACCTAATGGCTACTTCTGTGTGTATCAAGATCAAGAATCTTTTGAATACATCCGAGCTTGTGCCGAGAAGACTTTGCTTTGTAGTTTGGAACAAGAAGACAAACTGCCTTTTCAAGGTCTGCCGCTAGTCGTGATGTTCGTACAAGACGAGGGAATGGATAAGAAGGAGCTGGCCAGACTCCAAGAGGAGGGTCAGAATCTCGCCGACAACCTCCACTGCTCGTATATGGAGGCGTCCATCGGCGAGCTGGGCACGGAGGCGCTGACCCGCGAGGCGGTGCAGGAGCTGGTGCGCGCCAACAGGGACAAGGCGAGCTACGCGCATTTGTACCGTGACGTCATTGTGTGCTTCGACTCCGATATCAGAATAATGGTGTGCATGTTCTGCGACGACCCGTACTCGGCGGAGCGAGTGCTGAGTCCGTTCCTGTCGCACCGCGGCTGCTTCCTCACCGGCGACCGCTCGCTGCTCATAGAGACCTTCCTCGGAGACTCCAAACGGAAGGTCGAGGTAATAATATCAAGTTTCCACGGAGCCAGTCAATTCAGAGAGGAGCTGATACACGGTTTCATTCTGATTTATTCGGCGAAGAGAAAAGCCAGCTTAGCTACCTTAAATGCGTTTTCGATGAACATACCAAACCTGCCAATACAAATGGTGGCGGTGACGGAGGGCGGCGGCTCGGCGGCCAGCGCGTTCTTCGGCACGGACGTCGGGCACGCGCTGCTCACGGACGGGAACGCCACCGCCGACCGCCTCGCCGCGCACTTCGCCACCTACTCCTGCAGCGCAGACGCCAAATCCGCGTTCTACACGTCGTTCTTCAAGGAGGTGTGGGAGCGCAAGGGCGAGATCGAGCGCGCCTTCCGCCTGGAGCAGCCCGGCGCCGCGCACAACCTGCCCGACGTGGCCGCGGCGCGCGCGCCCGCTCGCTGCGACCCCGCGAGGGAGCACAAGTTAACGAACTCGTTGGATCTGCTCATCGGGCCCGACTCGAGAGCCGACATCGACTCTGAGTACAGCGACACCCTGAACAACTCCAAGAACAGAGGATTCTTGAAAGGTTTCAGCGTGTACCCCCCTCCCAGCACGCCGCCGGAGCCTGCGCCCCCGGACCACAGAATGCACCCGGATTTGTCTCCGGATCTCAACTGTTCTGAAGATTCACTGAGCACACACGAGTCAGATGGCGGTGGGTTGTGGCAGCCTCCCGGCTACGGGCACCGCGCCTTTACCACGGGACGCACGCGCCCGCACCCGCCGCCCCCGCGCCCCCGACACTCACAGACGCTCAAGCAACCCGGTAAATTGGACATGAACAACTACACGATGGTCAGCGACGCTCTACAACACATCACTATAGGTCCCCCGGCTCACTCCCGAGACAGGAAGTCTCAACGTTCCGGCTGGTCACACTCGAGCGGTCAGCAGGGTCACCACCACCACCCCTCCGGAGTCAGCTCGGAAACTGAACTGGATGCTCAATACGCGCAGATTAAAGAAACGAACGAGTATATGGAGGCGCCATCTATGACGCGCTTGAGGAGACATCGGAGACACGAGAAGACACCGCTGCACCAACCATCGTTCTCGGAGACGGACAGTTCCGGTTCTAGCGAAGCGTCCGGTGCCGGACGCGCGCCCGCCCGCCGCCGCCACGCCGCGCACCACGCGCCGCGACACTACAAGAAACGGTCCTTAGGTAACCTGGTGGCGGTGCAAAGCCCGCGAGTCCCGAAGCTCGGCATGTTCGTGGGGCCGCCGGAGCTGCCCACCGGCTACAGAGGTAGAACGCAAGAGGACAAGGCCGCCAGCGAGTCTAGCGACGGCAGCTCGGACGCGGACACGCGGCGGGTGCGCCCGCTGCCGCCGCCCCCGCACCACGAGCCCACCCACAAGATACTTGCCGGCGAGTACCCGGCGTCGATACAGGAGAACTGTACGGCCAGCGCGCCGGACCCGCGCCGCAGACCTCACCCCTTCGGCAAACACGACAGGAGGCACAAGGAATTCTCGAAGAGTAAGGACAAGAAGAACTCATCACAAGCAGCCAATGCAGCGTCAATTCAGAATTGGGGTCCGCAGGGCGGACACGGCGTTCCACTGTTCGTAGAGAAGTGCATAGAGTTCATAGAGCGCGAAGGGCTGGGCTCCGAGGGACTGTACCGCGTGCCTGGAAATAGGGCTCACGTCGATATGCTCTTCACCAAGTTCTACGAAGATCCGAATATAGATTTAGATTCGCTAGACATACCCGTGAATGCTGTCGCCACTGCGCTCAAGGATTTCTTCTCGAAGAAATTACCGCCATTACTCGATGAGGCGAGCATGGCGCAATTGGAAGATATCGCGAAAGTGCACGTTGTGGGTACAGCGATGCGCGGCTGCATGGCGGGCGGCGTGGAGCTCAAGGACCGCAGCTGGCGGCTGCTGGCCCTGCGCGCGCTGCTGCACTCCGCGCTCACGCCGCTCGCGCGCGCCACGCTCGACTACTTGCTGCACCACTTCGCCAGATTGGATAGAATCTCTATAAACTCCTTAAAATGTTCATAA
Protein
MAKKSDNTGGKLVTISVVGLSGTEKEKGQLGVGKSCLCNRFVRTHADDYNVDHICVLSQSDFSGRVVNNDHFLYWGNVQKENDDQEYRFEIVEQTEFVDDACFQPFKVGKTEPYVKRCVATKLQSAEKYMYVCKNQLGIEKEYEQKLMPEGKLTVDGFMCVYDVSLVPGRTWEKQNEIIAAILQNIIKLKKPVVLVTTKNDEACDQGVREAERLVQRKEFKGSIPIVETSSHDNVNVDQAFFLLAQMIDKSKARIKVANYAEALRIRRETLDFVTEAFTQLIRIHVEDHKELWSSVSKRLCHHAEWIKFVQQFGNDGTQVVFRRHVRRLKEERSAKKLRNQLAKLPQVLARLQLPTEELQENDWPVVVRQLRLHRDFSVYFSEGRGADSCSETGSELDSPLAIDTTLRVGERTRYQTLGQSQKIPYEILETNEAAAVFKSFLHEAQEEQRNYEWCQQFKRLLEETGYVTPGKQLSEVRVLLMGRECYEALSEEQQQRVYDQHQRQIQRRAKHNFQELLLEHADLFYHFKSISPTGTITQEDIKEITDVLQDDFRYKMLDRMEQDRKLMLFQHLGFVHCPMREHCPAGPNCLDATLPVILNTRVGSLTSAGESQCLAGPPAAPWALTTDTNHINVIILGMEGVAGEFGKRLMAGCDSERRINVHGHSWRLEQRIRTDDFSETTSIDDFAPNGYFCVYQDQESFEYIRACAEKTLLCSLEQEDKLPFQGLPLVVMFVQDEGMDKKELARLQEEGQNLADNLHCSYMEASIGELGTEALTREAVQELVRANRDKASYAHLYRDVIVCFDSDIRIMVCMFCDDPYSAERVLSPFLSHRGCFLTGDRSLLIETFLGDSKRKVEVIISSFHGASQFREELIHGFILIYSAKRKASLATLNAFSMNIPNLPIQMVAVTEGGGSAASAFFGTDVGHALLTDGNATADRLAAHFATYSCSADAKSAFYTSFFKEVWERKGEIERAFRLEQPGAAHNLPDVAAARAPARCDPAREHKLTNSLDLLIGPDSRADIDSEYSDTLNNSKNRGFLKGFSVYPPPSTPPEPAPPDHRMHPDLSPDLNCSEDSLSTHESDGGGLWQPPGYGHRAFTTGRTRPHPPPPRPRHSQTLKQPGKLDMNNYTMVSDALQHITIGPPAHSRDRKSQRSGWSHSSGQQGHHHHPSGVSSETELDAQYAQIKETNEYMEAPSMTRLRRHRRHEKTPLHQPSFSETDSSGSSEASGAGRAPARRRHAAHHAPRHYKKRSLGNLVAVQSPRVPKLGMFVGPPELPTGYRGRTQEDKAASESSDGSSDADTRRVRPLPPPPHHEPTHKILAGEYPASIQENCTASAPDPRRRPHPFGKHDRRHKEFSKSKDKKNSSQAANAASIQNWGPQGGHGVPLFVEKCIEFIEREGLGSEGLYRVPGNRAHVDMLFTKFYEDPNIDLDSLDIPVNAVATALKDFFSKKLPPLLDEASMAQLEDIAKVHVVGTAMRGCMAGGVELKDRSWRLLALRALLHSALTPLARATLDYLLHHFARLDRISINSLKCS
Summary
Uniprot
H9JV60
A0A3S2TRZ6
A0A2H1V6C9
A0A194PVW5
A0A1Y1KQ67
A0A1Y1KQ54
+ More
A0A1Y1KQ68 A0A1Y1KTM3 A0A1Y1KSJ7 A0A1Y1KSI6 A0A139WPM0 D6W848 A0A139WPB0 A0A139WPL6 A0A139WPT7 A0A139WPB3 E2C5J7 A0A0J7L0Y0 A0A1B6D9I5 A0A310SAT8 F4WQS4 E2A9N1 A0A195CZU1 A0A195EG12 A0A1B6DTL1 A0A195BRL7 A0A1B6C2R3 A0A026WFE0 A0A195ERU0 N6TE97 A0A232EWD7 A0A1B6K9C5 A0A1B6LPU4 A0A2S2QI88 E0VLS6 J9K7E6 A0A2S2P1B2 A0A0P6J4T1 A0A182GV26 A0A1Q3G0P2 A0A1Q3FVT6
A0A1Y1KQ68 A0A1Y1KTM3 A0A1Y1KSJ7 A0A1Y1KSI6 A0A139WPM0 D6W848 A0A139WPB0 A0A139WPL6 A0A139WPT7 A0A139WPB3 E2C5J7 A0A0J7L0Y0 A0A1B6D9I5 A0A310SAT8 F4WQS4 E2A9N1 A0A195CZU1 A0A195EG12 A0A1B6DTL1 A0A195BRL7 A0A1B6C2R3 A0A026WFE0 A0A195ERU0 N6TE97 A0A232EWD7 A0A1B6K9C5 A0A1B6LPU4 A0A2S2QI88 E0VLS6 J9K7E6 A0A2S2P1B2 A0A0P6J4T1 A0A182GV26 A0A1Q3G0P2 A0A1Q3FVT6
Pubmed
EMBL
BABH01026459
BABH01026460
BABH01026461
BABH01026462
BABH01026463
RSAL01000012
+ More
RVE53521.1 ODYU01000720 SOQ35952.1 KQ459590 KPI97467.1 GEZM01076910 JAV63559.1 GEZM01076916 JAV63549.1 GEZM01076914 JAV63552.1 GEZM01076909 JAV63560.1 GEZM01076913 GEZM01076911 JAV63558.1 GEZM01076917 JAV63548.1 KQ971307 KYB29836.1 EFA10998.2 KYB29839.1 KYB29837.1 KYB29835.1 KYB29838.1 GL452770 EFN76810.1 LBMM01001449 KMQ96291.1 GEDC01014951 JAS22347.1 KQ778189 OAD52014.1 GL888275 EGI63437.1 GL437918 EFN69833.1 KQ977041 KYN06136.1 KQ978983 KYN26834.1 GEDC01008319 JAS28979.1 KQ976424 KYM88586.1 GEDC01029497 JAS07801.1 KK107274 EZA53739.1 KQ981993 KYN30896.1 APGK01032712 KB740848 KB632411 ENN78679.1 ERL95234.1 NNAY01001878 OXU22660.1 GEBQ01031930 JAT08047.1 GEBQ01014443 JAT25534.1 GGMS01008218 MBY77421.1 DS235281 EEB14332.1 ABLF02022299 ABLF02022300 GGMR01010576 MBY23195.1 GDUN01000811 JAN95108.1 JXUM01090180 JXUM01090181 JXUM01090182 JXUM01090183 JXUM01090184 JXUM01090185 KQ563823 KXJ73225.1 GFDL01001677 JAV33368.1 GFDL01003348 JAV31697.1
RVE53521.1 ODYU01000720 SOQ35952.1 KQ459590 KPI97467.1 GEZM01076910 JAV63559.1 GEZM01076916 JAV63549.1 GEZM01076914 JAV63552.1 GEZM01076909 JAV63560.1 GEZM01076913 GEZM01076911 JAV63558.1 GEZM01076917 JAV63548.1 KQ971307 KYB29836.1 EFA10998.2 KYB29839.1 KYB29837.1 KYB29835.1 KYB29838.1 GL452770 EFN76810.1 LBMM01001449 KMQ96291.1 GEDC01014951 JAS22347.1 KQ778189 OAD52014.1 GL888275 EGI63437.1 GL437918 EFN69833.1 KQ977041 KYN06136.1 KQ978983 KYN26834.1 GEDC01008319 JAS28979.1 KQ976424 KYM88586.1 GEDC01029497 JAS07801.1 KK107274 EZA53739.1 KQ981993 KYN30896.1 APGK01032712 KB740848 KB632411 ENN78679.1 ERL95234.1 NNAY01001878 OXU22660.1 GEBQ01031930 JAT08047.1 GEBQ01014443 JAT25534.1 GGMS01008218 MBY77421.1 DS235281 EEB14332.1 ABLF02022299 ABLF02022300 GGMR01010576 MBY23195.1 GDUN01000811 JAN95108.1 JXUM01090180 JXUM01090181 JXUM01090182 JXUM01090183 JXUM01090184 JXUM01090185 KQ563823 KXJ73225.1 GFDL01001677 JAV33368.1 GFDL01003348 JAV31697.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JV60
A0A3S2TRZ6
A0A2H1V6C9
A0A194PVW5
A0A1Y1KQ67
A0A1Y1KQ54
+ More
A0A1Y1KQ68 A0A1Y1KTM3 A0A1Y1KSJ7 A0A1Y1KSI6 A0A139WPM0 D6W848 A0A139WPB0 A0A139WPL6 A0A139WPT7 A0A139WPB3 E2C5J7 A0A0J7L0Y0 A0A1B6D9I5 A0A310SAT8 F4WQS4 E2A9N1 A0A195CZU1 A0A195EG12 A0A1B6DTL1 A0A195BRL7 A0A1B6C2R3 A0A026WFE0 A0A195ERU0 N6TE97 A0A232EWD7 A0A1B6K9C5 A0A1B6LPU4 A0A2S2QI88 E0VLS6 J9K7E6 A0A2S2P1B2 A0A0P6J4T1 A0A182GV26 A0A1Q3G0P2 A0A1Q3FVT6
A0A1Y1KQ68 A0A1Y1KTM3 A0A1Y1KSJ7 A0A1Y1KSI6 A0A139WPM0 D6W848 A0A139WPB0 A0A139WPL6 A0A139WPT7 A0A139WPB3 E2C5J7 A0A0J7L0Y0 A0A1B6D9I5 A0A310SAT8 F4WQS4 E2A9N1 A0A195CZU1 A0A195EG12 A0A1B6DTL1 A0A195BRL7 A0A1B6C2R3 A0A026WFE0 A0A195ERU0 N6TE97 A0A232EWD7 A0A1B6K9C5 A0A1B6LPU4 A0A2S2QI88 E0VLS6 J9K7E6 A0A2S2P1B2 A0A0P6J4T1 A0A182GV26 A0A1Q3G0P2 A0A1Q3FVT6
PDB
6D4G
E-value=7.06835e-67,
Score=650
Ontologies
GO
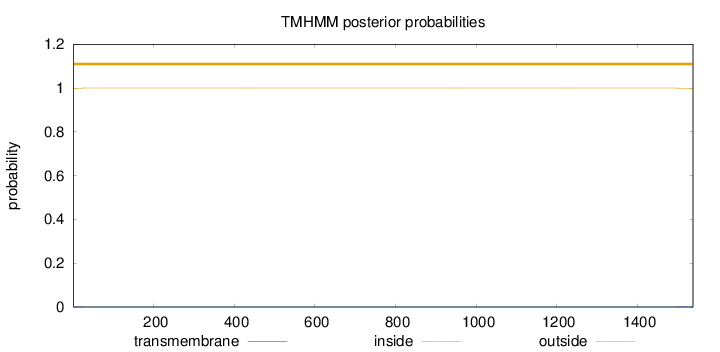
Topology
Length:
1538
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02849
Exp number, first 60 AAs:
0.00692
Total prob of N-in:
0.00041
outside
1 - 1538
Population Genetic Test Statistics
Pi
229.454927
Theta
170.956273
Tajima's D
1.196159
CLR
0.141623
CSRT
0.71636418179091
Interpretation
Uncertain
