Gene
KWMTBOMO15945
Pre Gene Modal
BGIBMGA013422
Annotation
hypothetical_protein_RR46_01282_[Papilio_xuthus]
Full name
tRNA-dihydrouridine(47) synthase [NAD(P)(+)]
+ More
Tyrosine-protein kinase
Tyrosine-protein kinase
Alternative Name
tRNA-dihydrouridine synthase 3
Location in the cell
Nuclear Reliability : 4.265
Sequence
CDS
ATGAAAGAATTTAAATACGATGTAGTTACTCTTATTCAAAGTGTACGAGATCGTCCCTGTTTATGGGATAAAACTTTAGAAAGCTACAAAGATCGAGTCGAGAGAAAAGCAGCGTGGGAGGAAATTTATAACATACTCGACGATAGATACGAGGAAATGACGCCTGTAGAAAAAAGGTTAACGGAGGAACACGTCGTCAACAAATGGACGAACGTTCGAGATACATTTGTCAAAACATTGAAGACGAAAATGGGAAGACCGAAACGCAAATACGTCTTTCACGAACATTTGAAGTTCCTTACAAAAGGAGCGCCCGATGAAACAGCTGAATACAGTGAATTTAGATCTGGCTTAATGAAACAAGAAGACGAATCGATTTTAGACAGCAGTAGCCAGTGTTACGACAAAAGCGATTCTATTTACACGCCAAAGAAGAAAAGAAAACGCGAGTCGGATAGTTATTCGATTGAAGAATACAATTACAAGAGGGAATCTAAAAGCAAAGAAGATAAGCAGAGTCATGATGAAACTAATGACATAGATTTCGTTGAGGTGGATACGAACGATCCAAGAGTGATGAATGAAGATGAAGCCTTTTTTGCTTCACTCCTGCCGAGCGTTGTGAGGTATGATGAAGACGAACGCTTGGAATTCCGTATAGAAGTGTTGGCTGTGATGAAAAAGATCAAGGAGAAGAGAAAATGGACGAGAATCGACTAA
Protein
MKEFKYDVVTLIQSVRDRPCLWDKTLESYKDRVERKAAWEEIYNILDDRYEEMTPVEKRLTEEHVVNKWTNVRDTFVKTLKTKMGRPKRKYVFHEHLKFLTKGAPDETAEYSEFRSGLMKQEDESILDSSSQCYDKSDSIYTPKKKRKRESDSYSIEEYNYKRESKSKEDKQSHDETNDIDFVEVDTNDPRVMNEDEAFFASLLPSVVRYDEDERLEFRIEVLAVMKKIKEKRKWTRID
Summary
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Cofactor
FMN
Similarity
Belongs to the dus family. Dus3 subfamily.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Uniprot
H9JV59
A0A0N1ICP7
A0A0N1PGE3
A0A1E1WHR4
A0A2A4JF93
A0A212FB26
+ More
A0A2H1VV20 A0A0L7LV89 A0A0T6BB68 A0A1E1VYW4 A0A2H1VGY6 A0A182GBC7 A0A2A4K4Q0 A0A182H144 A0A2S2NVF4 A0A2W1BQ58 A0A1E1WMV5 A0A1I8NIT7 A0A0T6B6I9 A0A1B6HMR2 A0A0L7L5Y0 A0A0N0PCI2 A0A1B6ED40 A0A1B6FZ56 A0A0R3R2M1 H9IVN4 A0A0P4WIC9 A0A0H5S1L7 A0A2S2P7V0 A0A1B6HMN4 A0A0N5D5I8 A0A1B6MVI3 A0A2J7PWA5 A0A1B6HE62 A0A154PSQ0 A0A194Q0C4 A0A2A4K541 A0A0J7K819 A0A0N4TX01 A0A2L2YL32 J9EDY0 A0A0L7L3E3 A0A067R379 A0A0A9WD81 A0A3P7E1U8 H9JU57 A0A146MBD5 A0A067RDN8 A0A0A9WIZ1 A0A2J7PB92 A0A0L7KVT9 A0A044V964 A0A1W4WXY7 A0A3P6U100 A0A182EJB9 A0A1B6HFE0 A0A2S2QKM5 A0A158Q924 A0A2W1BRZ0 A0A1E1WA50 A0A0P4WQG1 A0A1B6JE76 A0A1B6IF93 A0A212EVC2 A0A2H1WP32 A0A2J7RMA7 A0A0J7L3L1 A0A2J7PBC8 A0A026WD33 A0A2H1VEM6 A0A2S2PS33 A0A2J7RM80 A0A0P4VZP6 T1HA02 A0A0P4W354 A0A0P4W9X1 A0A2A4J2G2 J9KU42 A0A212F8H6 A0A212FPD0 A0A194PEH4 A0A0N0BFW2 A0A1W4X7D2 A0A1B6JD87 A0A1E1WAP8 A0A154P6K1
A0A2H1VV20 A0A0L7LV89 A0A0T6BB68 A0A1E1VYW4 A0A2H1VGY6 A0A182GBC7 A0A2A4K4Q0 A0A182H144 A0A2S2NVF4 A0A2W1BQ58 A0A1E1WMV5 A0A1I8NIT7 A0A0T6B6I9 A0A1B6HMR2 A0A0L7L5Y0 A0A0N0PCI2 A0A1B6ED40 A0A1B6FZ56 A0A0R3R2M1 H9IVN4 A0A0P4WIC9 A0A0H5S1L7 A0A2S2P7V0 A0A1B6HMN4 A0A0N5D5I8 A0A1B6MVI3 A0A2J7PWA5 A0A1B6HE62 A0A154PSQ0 A0A194Q0C4 A0A2A4K541 A0A0J7K819 A0A0N4TX01 A0A2L2YL32 J9EDY0 A0A0L7L3E3 A0A067R379 A0A0A9WD81 A0A3P7E1U8 H9JU57 A0A146MBD5 A0A067RDN8 A0A0A9WIZ1 A0A2J7PB92 A0A0L7KVT9 A0A044V964 A0A1W4WXY7 A0A3P6U100 A0A182EJB9 A0A1B6HFE0 A0A2S2QKM5 A0A158Q924 A0A2W1BRZ0 A0A1E1WA50 A0A0P4WQG1 A0A1B6JE76 A0A1B6IF93 A0A212EVC2 A0A2H1WP32 A0A2J7RMA7 A0A0J7L3L1 A0A2J7PBC8 A0A026WD33 A0A2H1VEM6 A0A2S2PS33 A0A2J7RM80 A0A0P4VZP6 T1HA02 A0A0P4W354 A0A0P4W9X1 A0A2A4J2G2 J9KU42 A0A212F8H6 A0A212FPD0 A0A194PEH4 A0A0N0BFW2 A0A1W4X7D2 A0A1B6JD87 A0A1E1WAP8 A0A154P6K1
EC Number
1.3.1.-
2.7.10.2
2.7.10.2
Pubmed
EMBL
BABH01026450
KQ459572
KPI99593.1
KQ460448
KPJ14681.1
GDQN01004532
+ More
JAT86522.1 NWSH01001707 PCG70378.1 AGBW02009394 OWR50923.1 ODYU01004606 SOQ44670.1 JTDY01000013 KOB79428.1 LJIG01002364 KRT84576.1 GDQN01011180 JAT79874.1 ODYU01002266 SOQ39524.1 JXUM01052702 KQ561745 KXJ77651.1 NWSH01000176 PCG78743.1 JXUM01102437 KQ564727 KXJ71823.1 GGMR01008582 MBY21201.1 KZ149935 PZC77192.1 GDQN01002878 JAT88176.1 LJIG01009493 KRT82969.1 GECU01031770 JAS75936.1 JTDY01002813 KOB70669.1 KQ460651 KPJ13045.1 GEDC01001514 JAS35784.1 GECZ01014282 JAS55487.1 UZAG01019009 VDO41994.1 BABH01039610 GDRN01070949 JAI63786.1 LN856534 CRZ22603.1 GGMR01012912 MBY25531.1 GECU01031767 JAS75939.1 UYYF01004606 VDN05797.1 GEBQ01000068 JAT39909.1 NEVH01020936 PNF20615.1 GECU01034815 JAS72891.1 KQ435163 KZC14931.1 KQ459582 KPI99032.1 NWSH01000149 PCG79034.1 LBMM01011903 KMQ86558.1 UZAD01013389 VDN94591.1 IAAA01038977 LAA08833.1 ADBV01004300 EJW80661.1 JTDY01003266 KOB69841.1 KK852738 KDR17414.1 GBHO01037870 JAG05734.1 UYWW01001533 VDM10647.1 BABH01004110 GDHC01001538 JAQ17091.1 KK852767 KDR16963.1 GBHO01035202 JAG08402.1 NEVH01027172 PNF13604.1 JTDY01005074 KOB67397.1 CMVM020000255 UYRW01003313 VDK88653.1 GECU01034306 JAS73400.1 GGMS01009116 MBY78319.1 KZ149960 PZC76385.1 GDQN01007182 JAT83872.1 GDRN01070950 JAI63785.1 GECU01010140 JAS97566.1 GECU01022098 JAS85608.1 AGBW02012210 OWR45442.1 ODYU01010020 SOQ54829.1 NEVH01002568 PNF41952.1 LBMM01000937 KMQ97188.1 NEVH01027118 NEVH01016302 NEVH01007440 PNF13628.1 PNF25815.1 PNF35690.1 KK107261 QOIP01000012 EZA53972.1 RLU16330.1 ODYU01002127 SOQ39226.1 GGMR01019594 MBY32213.1 PNF41936.1 GDKW01001597 JAI54998.1 ACPB03005729 GDRN01106643 JAI57541.1 GDRN01106646 GDRN01106645 JAI57540.1 NWSH01003900 PCG65714.1 ABLF02030014 AGBW02009720 OWR50045.1 AGBW02003328 OWR55560.1 KQ459606 KPI91756.1 KQ435794 KOX73672.1 GECU01010480 JAS97226.1 GDQN01006974 JAT84080.1 KQ434827 KZC07471.1
JAT86522.1 NWSH01001707 PCG70378.1 AGBW02009394 OWR50923.1 ODYU01004606 SOQ44670.1 JTDY01000013 KOB79428.1 LJIG01002364 KRT84576.1 GDQN01011180 JAT79874.1 ODYU01002266 SOQ39524.1 JXUM01052702 KQ561745 KXJ77651.1 NWSH01000176 PCG78743.1 JXUM01102437 KQ564727 KXJ71823.1 GGMR01008582 MBY21201.1 KZ149935 PZC77192.1 GDQN01002878 JAT88176.1 LJIG01009493 KRT82969.1 GECU01031770 JAS75936.1 JTDY01002813 KOB70669.1 KQ460651 KPJ13045.1 GEDC01001514 JAS35784.1 GECZ01014282 JAS55487.1 UZAG01019009 VDO41994.1 BABH01039610 GDRN01070949 JAI63786.1 LN856534 CRZ22603.1 GGMR01012912 MBY25531.1 GECU01031767 JAS75939.1 UYYF01004606 VDN05797.1 GEBQ01000068 JAT39909.1 NEVH01020936 PNF20615.1 GECU01034815 JAS72891.1 KQ435163 KZC14931.1 KQ459582 KPI99032.1 NWSH01000149 PCG79034.1 LBMM01011903 KMQ86558.1 UZAD01013389 VDN94591.1 IAAA01038977 LAA08833.1 ADBV01004300 EJW80661.1 JTDY01003266 KOB69841.1 KK852738 KDR17414.1 GBHO01037870 JAG05734.1 UYWW01001533 VDM10647.1 BABH01004110 GDHC01001538 JAQ17091.1 KK852767 KDR16963.1 GBHO01035202 JAG08402.1 NEVH01027172 PNF13604.1 JTDY01005074 KOB67397.1 CMVM020000255 UYRW01003313 VDK88653.1 GECU01034306 JAS73400.1 GGMS01009116 MBY78319.1 KZ149960 PZC76385.1 GDQN01007182 JAT83872.1 GDRN01070950 JAI63785.1 GECU01010140 JAS97566.1 GECU01022098 JAS85608.1 AGBW02012210 OWR45442.1 ODYU01010020 SOQ54829.1 NEVH01002568 PNF41952.1 LBMM01000937 KMQ97188.1 NEVH01027118 NEVH01016302 NEVH01007440 PNF13628.1 PNF25815.1 PNF35690.1 KK107261 QOIP01000012 EZA53972.1 RLU16330.1 ODYU01002127 SOQ39226.1 GGMR01019594 MBY32213.1 PNF41936.1 GDKW01001597 JAI54998.1 ACPB03005729 GDRN01106643 JAI57541.1 GDRN01106646 GDRN01106645 JAI57540.1 NWSH01003900 PCG65714.1 ABLF02030014 AGBW02009720 OWR50045.1 AGBW02003328 OWR55560.1 KQ459606 KPI91756.1 KQ435794 KOX73672.1 GECU01010480 JAS97226.1 GDQN01006974 JAT84080.1 KQ434827 KZC07471.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000037510
+ More
UP000069940 UP000249989 UP000095301 UP000050602 UP000006672 UP000046394 UP000276776 UP000235965 UP000076502 UP000036403 UP000038020 UP000278627 UP000004810 UP000093561 UP000027135 UP000270924 UP000024404 UP000192223 UP000271087 UP000077448 UP000050640 UP000053097 UP000279307 UP000015103 UP000007819 UP000053105
UP000069940 UP000249989 UP000095301 UP000050602 UP000006672 UP000046394 UP000276776 UP000235965 UP000076502 UP000036403 UP000038020 UP000278627 UP000004810 UP000093561 UP000027135 UP000270924 UP000024404 UP000192223 UP000271087 UP000077448 UP000050640 UP000053097 UP000279307 UP000015103 UP000007819 UP000053105
Pfam
Interpro
IPR006578
MADF-dom
+ More
IPR039353 TF_Adf1
IPR004210 BESS_motif
IPR000753 Clusterin-like
IPR013785 Aldolase_TIM
IPR000571 Znf_CCCH
IPR018517 tRNA_hU_synthase_CS
IPR001269 tRNA_hU_synthase
IPR035587 DUS-like_FMN-bd
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR036028 SH3-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000980 SH2
IPR000719 Prot_kinase_dom
IPR001452 SH3_domain
IPR020635 Tyr_kinase_cat_dom
IPR036860 SH2_dom_sf
IPR008266 Tyr_kinase_AS
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR018247 EF_Hand_1_Ca_BS
IPR039353 TF_Adf1
IPR004210 BESS_motif
IPR000753 Clusterin-like
IPR013785 Aldolase_TIM
IPR000571 Znf_CCCH
IPR018517 tRNA_hU_synthase_CS
IPR001269 tRNA_hU_synthase
IPR035587 DUS-like_FMN-bd
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR036028 SH3-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000980 SH2
IPR000719 Prot_kinase_dom
IPR001452 SH3_domain
IPR020635 Tyr_kinase_cat_dom
IPR036860 SH2_dom_sf
IPR008266 Tyr_kinase_AS
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR018247 EF_Hand_1_Ca_BS
Gene 3D
ProteinModelPortal
H9JV59
A0A0N1ICP7
A0A0N1PGE3
A0A1E1WHR4
A0A2A4JF93
A0A212FB26
+ More
A0A2H1VV20 A0A0L7LV89 A0A0T6BB68 A0A1E1VYW4 A0A2H1VGY6 A0A182GBC7 A0A2A4K4Q0 A0A182H144 A0A2S2NVF4 A0A2W1BQ58 A0A1E1WMV5 A0A1I8NIT7 A0A0T6B6I9 A0A1B6HMR2 A0A0L7L5Y0 A0A0N0PCI2 A0A1B6ED40 A0A1B6FZ56 A0A0R3R2M1 H9IVN4 A0A0P4WIC9 A0A0H5S1L7 A0A2S2P7V0 A0A1B6HMN4 A0A0N5D5I8 A0A1B6MVI3 A0A2J7PWA5 A0A1B6HE62 A0A154PSQ0 A0A194Q0C4 A0A2A4K541 A0A0J7K819 A0A0N4TX01 A0A2L2YL32 J9EDY0 A0A0L7L3E3 A0A067R379 A0A0A9WD81 A0A3P7E1U8 H9JU57 A0A146MBD5 A0A067RDN8 A0A0A9WIZ1 A0A2J7PB92 A0A0L7KVT9 A0A044V964 A0A1W4WXY7 A0A3P6U100 A0A182EJB9 A0A1B6HFE0 A0A2S2QKM5 A0A158Q924 A0A2W1BRZ0 A0A1E1WA50 A0A0P4WQG1 A0A1B6JE76 A0A1B6IF93 A0A212EVC2 A0A2H1WP32 A0A2J7RMA7 A0A0J7L3L1 A0A2J7PBC8 A0A026WD33 A0A2H1VEM6 A0A2S2PS33 A0A2J7RM80 A0A0P4VZP6 T1HA02 A0A0P4W354 A0A0P4W9X1 A0A2A4J2G2 J9KU42 A0A212F8H6 A0A212FPD0 A0A194PEH4 A0A0N0BFW2 A0A1W4X7D2 A0A1B6JD87 A0A1E1WAP8 A0A154P6K1
A0A2H1VV20 A0A0L7LV89 A0A0T6BB68 A0A1E1VYW4 A0A2H1VGY6 A0A182GBC7 A0A2A4K4Q0 A0A182H144 A0A2S2NVF4 A0A2W1BQ58 A0A1E1WMV5 A0A1I8NIT7 A0A0T6B6I9 A0A1B6HMR2 A0A0L7L5Y0 A0A0N0PCI2 A0A1B6ED40 A0A1B6FZ56 A0A0R3R2M1 H9IVN4 A0A0P4WIC9 A0A0H5S1L7 A0A2S2P7V0 A0A1B6HMN4 A0A0N5D5I8 A0A1B6MVI3 A0A2J7PWA5 A0A1B6HE62 A0A154PSQ0 A0A194Q0C4 A0A2A4K541 A0A0J7K819 A0A0N4TX01 A0A2L2YL32 J9EDY0 A0A0L7L3E3 A0A067R379 A0A0A9WD81 A0A3P7E1U8 H9JU57 A0A146MBD5 A0A067RDN8 A0A0A9WIZ1 A0A2J7PB92 A0A0L7KVT9 A0A044V964 A0A1W4WXY7 A0A3P6U100 A0A182EJB9 A0A1B6HFE0 A0A2S2QKM5 A0A158Q924 A0A2W1BRZ0 A0A1E1WA50 A0A0P4WQG1 A0A1B6JE76 A0A1B6IF93 A0A212EVC2 A0A2H1WP32 A0A2J7RMA7 A0A0J7L3L1 A0A2J7PBC8 A0A026WD33 A0A2H1VEM6 A0A2S2PS33 A0A2J7RM80 A0A0P4VZP6 T1HA02 A0A0P4W354 A0A0P4W9X1 A0A2A4J2G2 J9KU42 A0A212F8H6 A0A212FPD0 A0A194PEH4 A0A0N0BFW2 A0A1W4X7D2 A0A1B6JD87 A0A1E1WAP8 A0A154P6K1
Ontologies
GO
PANTHER
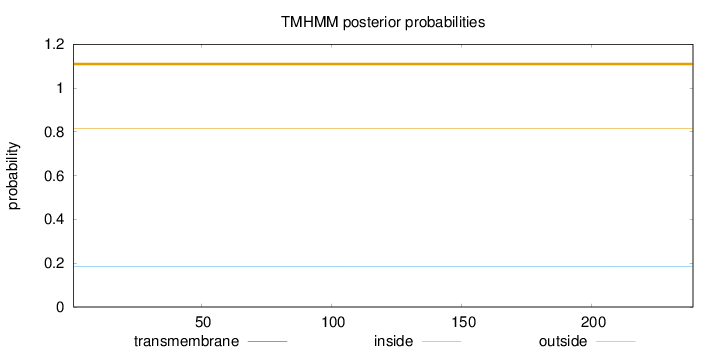
Topology
Subcellular location
Nucleus
Length:
239
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.18501
outside
1 - 239
Population Genetic Test Statistics
Pi
270.59977
Theta
187.877472
Tajima's D
1.362381
CLR
0.221926
CSRT
0.758762061896905
Interpretation
Uncertain
