Gene
KWMTBOMO15943
Pre Gene Modal
BGIBMGA013421
Annotation
vrille_[Antheraea_pernyi]
Transcription factor
Location in the cell
Nuclear Reliability : 4.25
Sequence
CDS
ATGGTTGCCGAGTTCATCCTGAGCCAGCAACAGCTGCTGAGCGCAGGGGCCGGTGCAGCCCTAGGGCCCGCCCCAGTGCAGACCCGCGCTCCTCCACCACCGCGCGCTCGTCTCTCCGTCTCGCCGCATTTCCCGATGGACCAAGCCCCGCCTTCCAACGGCAGCAGCGGCGACAGCCACGACTACGGAGCCTTCGACTTCAAACGCAAAGACTTCTTCGGGCAACGCAAACAACGCGAGTTCATACCGGACAGCAAAAAGGACGACGGCTACTGGGACCGCAGGAGGCGCAACAACGAGGCAGCCAAGCGGTCTCGCGAAAAACGCCGCTTCAACGACATGGTCCTGGAACAGCGCGTCGTCGAACTCTCGAAGGAAAACCACGTCCTCAAAGCACAATTAGACGCCATAAAAGAAAAATACGGAATATGTGGAGAAAGCTTGATAAGCATAGACCAAGTCTTAGCCACCTTGCCAACGTGTGATCAAGTTCTATGTGTGACAAAGAGGAGTAAACTTTCAGGCACAGCCTTGTTTCCGGCCGCTCCTCCTCCGCCCCCGCCGGCTCCACCAGCTTCTCCCCCGTCTCCCCCACCACAACGCGCCCCAGAGCCATACCAAGAAAGGATTCCCGTACCCGAAGCATACTACCCCCACCCTGCCCACTACGAACCACCCGGTTCAGTTCTCAACTTGTCCAGATCTCGACGTGCCCCGTCTCCTTATGAGCTATCTTCCTTATCTGGATCCGGAGACGAGACCGCCGAGCAGTACGCCCCAGAAAACAACTGCCTTCCTTTGAAACTGCGCCACAAATCTCACCTCGGCGACAAGGACGTCGCTAGCGCTCTGCTATCCCTCCAGCACATAAAGCAGGAGCCGGGGCCCAGGTCGTCCCCCTCCTGGGACGGCGAGGGTTCTAGCGACGAGCGCGACTCCGGGATCTCACTAGGAGCAGAATACAGACCGCAACCGGAAAGGCATCCGGAGGAGGAGGACGCGCATTTGAAGGCTGAGCTGGCACGGCTGGCCACAGAGGTGGCGACGCTCAAGAACATGATGCACCAGAACAAAACCCGCGCGCACGAGCACTGA
Protein
MVAEFILSQQQLLSAGAGAALGPAPVQTRAPPPPRARLSVSPHFPMDQAPPSNGSSGDSHDYGAFDFKRKDFFGQRKQREFIPDSKKDDGYWDRRRRNNEAAKRSREKRRFNDMVLEQRVVELSKENHVLKAQLDAIKEKYGICGESLISIDQVLATLPTCDQVLCVTKRSKLSGTALFPAAPPPPPPAPPASPPSPPPQRAPEPYQERIPVPEAYYPHPAHYEPPGSVLNLSRSRRAPSPYELSSLSGSGDETAEQYAPENNCLPLKLRHKSHLGDKDVASALLSLQHIKQEPGPRSSPSWDGEGSSDERDSGISLGAEYRPQPERHPEEEDAHLKAELARLATEVATLKNMMHQNKTRAHEH
Summary
Uniprot
H9JV58
Q699T4
A0A0N1I856
A0A0N0P9B0
Q1XD36
A0A2A4JG24
+ More
A0A3S2LTK9 A0A2H1VPE1 A0A0L7LW91 A0A212FB12 A0A0B4SXC5 A0A0B4SXR8 A0A0B4SWZ8 A0A0B4SXJ3 A0A0B4SX02 A0A0B4SXR4 A0A0B4SXP9 A0A0B4SY49 A0A0B4SXL7 A0A0B4SXD7 U4UNX2 A0A1B6HEF2 A0A1B6KIK3 K7ING0 A0A310SJK2 A0A154P2D0 A0A0L7R5C1 A0A2A3ELX9 V9ICA8 V9IA22 A0A0J7L319 A0A158NSB1 A0A195CWT6 A0A0M8ZQP5 A0A088A183 N6TIM2 V5G6G3 T1H8P6 E0VHE1 A0A0T6B5M4 A0A026X373 A0A023FAB3 A0A067R9W9 T1GMH3 E2AHN2 E2BJP9
A0A3S2LTK9 A0A2H1VPE1 A0A0L7LW91 A0A212FB12 A0A0B4SXC5 A0A0B4SXR8 A0A0B4SWZ8 A0A0B4SXJ3 A0A0B4SX02 A0A0B4SXR4 A0A0B4SXP9 A0A0B4SY49 A0A0B4SXL7 A0A0B4SXD7 U4UNX2 A0A1B6HEF2 A0A1B6KIK3 K7ING0 A0A310SJK2 A0A154P2D0 A0A0L7R5C1 A0A2A3ELX9 V9ICA8 V9IA22 A0A0J7L319 A0A158NSB1 A0A195CWT6 A0A0M8ZQP5 A0A088A183 N6TIM2 V5G6G3 T1H8P6 E0VHE1 A0A0T6B5M4 A0A026X373 A0A023FAB3 A0A067R9W9 T1GMH3 E2AHN2 E2BJP9
Pubmed
EMBL
BABH01026441
AY526608
AAS92609.1
KQ460448
KPJ14679.1
KQ459572
+ More
KPI99591.1 AY576272 AAT86041.1 NWSH01001707 PCG70380.1 RSAL01000012 RVE53525.1 ODYU01003648 SOQ42667.1 JTDY01000013 KOB79426.1 AGBW02009394 OWR50921.1 KM675552 KM675649 AJB84657.1 AJB84682.1 KM675644 KM675645 AJB84678.1 KM675556 KM675640 KM675642 KM675643 AJB84660.1 AJB84676.1 KM675555 KM675557 AJB84659.1 KM675551 KM675648 AJB84656.1 AJB84681.1 KM675639 AJB84673.1 KM675554 AJB84658.1 KM675647 AJB84680.1 KM675646 AJB84679.1 KM675558 KM675637 AJB84662.1 AJB84692.1 KB632399 ERL94812.1 GECU01034725 JAS72981.1 GEBQ01028696 JAT11281.1 KQ764881 OAD54376.1 KQ434792 KZC05494.1 KQ414652 KOC66077.1 KZ288215 PBC32710.1 JR037778 AEY57964.1 JR037777 AEY57963.1 LBMM01000921 KMQ97212.1 ADTU01024726 KQ977171 KYN05131.1 KQ435908 KOX68987.1 APGK01024789 KB740562 ENN80284.1 GALX01002796 JAB65670.1 ACPB03022038 DS235169 EEB12797.1 LJIG01009740 KRT82475.1 KK107019 QOIP01000007 EZA62735.1 RLU20592.1 GBBI01000286 JAC18426.1 KK852791 KDR16467.1 CAQQ02150384 CAQQ02150385 GL439566 EFN67066.1 GL448604 EFN84107.1
KPI99591.1 AY576272 AAT86041.1 NWSH01001707 PCG70380.1 RSAL01000012 RVE53525.1 ODYU01003648 SOQ42667.1 JTDY01000013 KOB79426.1 AGBW02009394 OWR50921.1 KM675552 KM675649 AJB84657.1 AJB84682.1 KM675644 KM675645 AJB84678.1 KM675556 KM675640 KM675642 KM675643 AJB84660.1 AJB84676.1 KM675555 KM675557 AJB84659.1 KM675551 KM675648 AJB84656.1 AJB84681.1 KM675639 AJB84673.1 KM675554 AJB84658.1 KM675647 AJB84680.1 KM675646 AJB84679.1 KM675558 KM675637 AJB84662.1 AJB84692.1 KB632399 ERL94812.1 GECU01034725 JAS72981.1 GEBQ01028696 JAT11281.1 KQ764881 OAD54376.1 KQ434792 KZC05494.1 KQ414652 KOC66077.1 KZ288215 PBC32710.1 JR037778 AEY57964.1 JR037777 AEY57963.1 LBMM01000921 KMQ97212.1 ADTU01024726 KQ977171 KYN05131.1 KQ435908 KOX68987.1 APGK01024789 KB740562 ENN80284.1 GALX01002796 JAB65670.1 ACPB03022038 DS235169 EEB12797.1 LJIG01009740 KRT82475.1 KK107019 QOIP01000007 EZA62735.1 RLU20592.1 GBBI01000286 JAC18426.1 KK852791 KDR16467.1 CAQQ02150384 CAQQ02150385 GL439566 EFN67066.1 GL448604 EFN84107.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000283053
UP000037510
+ More
UP000007151 UP000030742 UP000002358 UP000076502 UP000053825 UP000242457 UP000036403 UP000005205 UP000078542 UP000053105 UP000005203 UP000019118 UP000015103 UP000009046 UP000053097 UP000279307 UP000027135 UP000015102 UP000000311 UP000008237
UP000007151 UP000030742 UP000002358 UP000076502 UP000053825 UP000242457 UP000036403 UP000005205 UP000078542 UP000053105 UP000005203 UP000019118 UP000015103 UP000009046 UP000053097 UP000279307 UP000027135 UP000015102 UP000000311 UP000008237
PRIDE
Interpro
ProteinModelPortal
H9JV58
Q699T4
A0A0N1I856
A0A0N0P9B0
Q1XD36
A0A2A4JG24
+ More
A0A3S2LTK9 A0A2H1VPE1 A0A0L7LW91 A0A212FB12 A0A0B4SXC5 A0A0B4SXR8 A0A0B4SWZ8 A0A0B4SXJ3 A0A0B4SX02 A0A0B4SXR4 A0A0B4SXP9 A0A0B4SY49 A0A0B4SXL7 A0A0B4SXD7 U4UNX2 A0A1B6HEF2 A0A1B6KIK3 K7ING0 A0A310SJK2 A0A154P2D0 A0A0L7R5C1 A0A2A3ELX9 V9ICA8 V9IA22 A0A0J7L319 A0A158NSB1 A0A195CWT6 A0A0M8ZQP5 A0A088A183 N6TIM2 V5G6G3 T1H8P6 E0VHE1 A0A0T6B5M4 A0A026X373 A0A023FAB3 A0A067R9W9 T1GMH3 E2AHN2 E2BJP9
A0A3S2LTK9 A0A2H1VPE1 A0A0L7LW91 A0A212FB12 A0A0B4SXC5 A0A0B4SXR8 A0A0B4SWZ8 A0A0B4SXJ3 A0A0B4SX02 A0A0B4SXR4 A0A0B4SXP9 A0A0B4SY49 A0A0B4SXL7 A0A0B4SXD7 U4UNX2 A0A1B6HEF2 A0A1B6KIK3 K7ING0 A0A310SJK2 A0A154P2D0 A0A0L7R5C1 A0A2A3ELX9 V9ICA8 V9IA22 A0A0J7L319 A0A158NSB1 A0A195CWT6 A0A0M8ZQP5 A0A088A183 N6TIM2 V5G6G3 T1H8P6 E0VHE1 A0A0T6B5M4 A0A026X373 A0A023FAB3 A0A067R9W9 T1GMH3 E2AHN2 E2BJP9
PDB
6MG3
E-value=0.0623477,
Score=84
Ontologies
GO
Topology
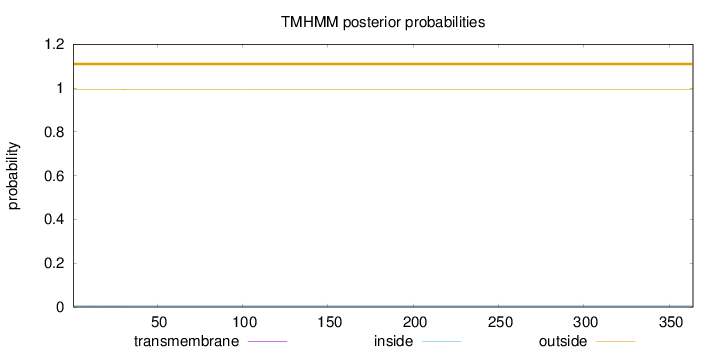
Length:
364
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02062
Exp number, first 60 AAs:
0.02062
Total prob of N-in:
0.00684
outside
1 - 364
Population Genetic Test Statistics
Pi
159.695369
Theta
177.384421
Tajima's D
-0.839401
CLR
339.417164
CSRT
0.167341632918354
Interpretation
Uncertain
