Gene
KWMTBOMO15942
Pre Gene Modal
BGIBMGA013382
Annotation
PREDICTED:_potassium_channel_subfamily_K_member_9-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.17
Sequence
CDS
ATGAAGAAGCAAAATGTACGCACTCTATCATTAATTGTGTGCACTTTCACATATTTGCTTGTCGGTGCAGCCGTTTTTGATGCACTTGAGTCGACGACCGAAAGGAACAGATTTGATGTGTTACAAGCGGTGATTGAAGCCATGATAATAAGAAAGTACAACATATCGGAGGAGGACTTCCTCATCATGGAGACAGTGGTGCTTAAATCAGAGCCCCACAAGGCTGGCCAGCAATGGAAGTTCACTGGTGCCTTTTACTATGCTACTACTGTGCTCACTACCATTGGCTATGGTCATTCAACGCCAGCTACGGTCAGCGGCAAGCTATTCACTATGTTCTATGCGATTGTTGGTATACCCTTGGGCTTGATTATGTTCCAAAGCATCGGTGAAAGAGTTAACAGGCTCAGCAGTGTAATAATAAAATCAATAAAACGAGCATTGAATTGCAAACAGACAAATGCAACGGAAGTGGATCTTATATGTGTTGTGACCACCCTGTCTTCCCTTACCATTGCTGGTGGTGCGGCCGCCTTCTCAACATTCGAGGGGTGGAGTTACTTTGATAGCGTTTACTATTGCTTCATAACTCTCACTACTATAGGTTTCGGTGACATGGTAGCCCTACAGAAAGACAACGCACTGAACTACAAGCCTTCTTATGTGATGTTTTCGCTAATTTTCATCCTGTTCGGGCTGGCTATAGTGGCGGCCTGCCTCAATTTGCTTGTTCTACGATTCGTTACTATGAACACTGAGGATGAAAAGAGAGACCAGCAGCAAGCAGAACAGGCTCAGCAAGTAGCGGTCCGGTTGGAGGGCGATGTGATCACTGCGGATGGCGACGTGTTGCGCGGCGTGTCGCGGGGCACCAGGCTGCCGGCTGACCCGAGGCAGATATCTACCGGTGACACATCCCTGATACCATTATACGTGGATGAAGACTACGCGCCCAGCGTCTGCAGTTGTTCGTGTAACTGCTTCGGACCGAACAGATCGATACCGTACCGAGATCCGCTATCGCCGGTGCCGCCCACGAACATAAAACAGATCAAATCGAAAAACTATCGAGACATCAGCGGCATACCGCGCTACATCGAGAAATCGTCATCGAAGAATCGTTCGCAATCGTTGCGATTAAACTCCGCATCCGGCTATCTCTACATGAATGCGAAAACGAACGATTTCCAACTCGAGGCCGAAGACTTCCGAGAGATCGTGGCGAAGAGACGCGAACAATTACGCAAAGGTATGATGATGTACGATATCCAATATAATAACGAGTATTTACAACCCTATCGACACAGTATTAGTACGCGGCTAGAGACGAGCCCGCACAGTTCGCTCTATTCGATGAACATTAGATCCGATAATCGATCGAACGATCCGCTTGTCGACGATTACGATTCCGAGAAATCGAGTTACGCGAAAAAGAAGCTTATGAAGAACATAGCAAGGAACACAAATAACAGACGAGTCGAGAATTATTTTTACGACGACGCCGTATTGCACTTCGACGAGGAATACGCGTTCGACGGTTCGATTTTGTTCGCGAAAAGGAGAAAATCGATCGATAAAAACTGGGAGGAGTTTGCGTGCGAATTGCCGAGGAGTTACGGCGCGGACCAAATATCGAACGATGGGAGCTACGGGTATTATTTGCCCGATGAAGACGAGCAAGAACAGTACTTTATAAACGACACGTTAACGTTCACTATGCCTCCGCACAGGGCGAGCATATAA
Protein
MKKQNVRTLSLIVCTFTYLLVGAAVFDALESTTERNRFDVLQAVIEAMIIRKYNISEEDFLIMETVVLKSEPHKAGQQWKFTGAFYYATTVLTTIGYGHSTPATVSGKLFTMFYAIVGIPLGLIMFQSIGERVNRLSSVIIKSIKRALNCKQTNATEVDLICVVTTLSSLTIAGGAAAFSTFEGWSYFDSVYYCFITLTTIGFGDMVALQKDNALNYKPSYVMFSLIFILFGLAIVAACLNLLVLRFVTMNTEDEKRDQQQAEQAQQVAVRLEGDVITADGDVLRGVSRGTRLPADPRQISTGDTSLIPLYVDEDYAPSVCSCSCNCFGPNRSIPYRDPLSPVPPTNIKQIKSKNYRDISGIPRYIEKSSSKNRSQSLRLNSASGYLYMNAKTNDFQLEAEDFREIVAKRREQLRKGMMMYDIQYNNEYLQPYRHSISTRLETSPHSSLYSMNIRSDNRSNDPLVDDYDSEKSSYAKKKLMKNIARNTNNRRVENYFYDDAVLHFDEEYAFDGSILFAKRRKSIDKNWEEFACELPRSYGADQISNDGSYGYYLPDEDEQEQYFINDTLTFTMPPHRASI
Summary
Similarity
Belongs to the two pore domain potassium channel (TC 1.A.1.8) family.
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF07885 Ion_trans_2
ProteinModelPortal
PDB
4XDL
E-value=2.11477e-27,
Score=306
Ontologies
GO
Topology
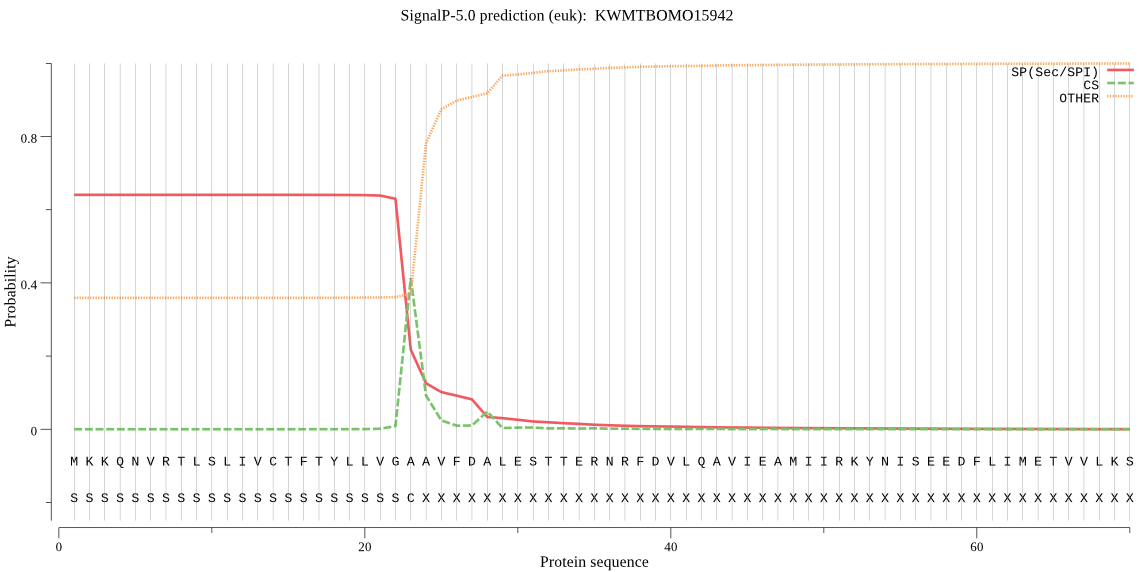
SignalP
Position: 1 - 23,
Likelihood: 0.640276
Length:
580
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
113.0784
Exp number, first 60 AAs:
19.21207
Total prob of N-in:
0.80532
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 108
TMhelix
109 - 126
inside
127 - 159
TMhelix
160 - 182
outside
183 - 186
TMhelix
187 - 209
inside
210 - 221
TMhelix
222 - 244
outside
245 - 580

Population Genetic Test Statistics
Pi
183.744486
Theta
143.028666
Tajima's D
0.767164
CLR
0.262568
CSRT
0.595070246487676
Interpretation
Uncertain
