Gene
KWMTBOMO15928 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003469
Annotation
poly_A_binding_protein_[Bombyx_mori]
Full name
Polyadenylate-binding protein
Location in the cell
Cytoplasmic Reliability : 2.158
Sequence
CDS
ATGAATCCTGGTCCACCAAGTTATCCGATGGCATCTCTGTATGTTGGAGATTTACACTCTGATATAACCGAGGCCATGTTGTTTGAAAAATTTTCGACTGCTGGGCCCGTCCTATCTATACGTGTTTGTCGCGATATGATTACCCGTAGATCCTTAGGCTACGCTTATGTTAATTTTCAGCAACCTACGGATGCTGAAAGAGCTTTAGACACCATGAACTTTGATATTATCAAAGGAAGACCCATTAGAATCATGTGGTCCCAACGTGATCCTTCCCTGCGCAAATCTGGAGTTGGGAATGTTTTTATAAAAAACCTTGATAAATCTATTGACAACAAGGCTTTATTTGATACATTCTCAGCGTTTGGGAATATTTTGAGTTGTAAAGTAGCTCAAGATGAGACTGGTGCTTCAAAAGGGTATGGTTTTGTTCATTTTGAAACTGAAGAAGCTGCAAATAAATCTATTGAAAAGGTAAATGGGATGTTGTTAAATGGTAAAATGGTTTATGTAGGCAGATTCATTCCACGCAAAGAGCGCGAAAAGGAACTTGGTGAGAAGGCTAAACTGTTTGCCAATGTGTATGTAAAGAACTTTGGAGAAGACTTTTCTGATGAAATGTTAAAAGATATGTTTGAGAAATATGGTCGCATTACAAGTCACAAAGTTATGTACAAAGATGATGGCAATTCTAGAGGCTTTGGTTTTGTTGCTTTTGAAGACCCAGATGCTGCTGAAAGAGCTTCCATTGAATTGAATGGCAAAGAGCTAGTTGAGGGTAAGCCTCTCTATGTTGGACGTGCACAAAAGAAAGCTGAACGCCAAAAGGAGTTGAAGCGTAAGTTTGAACAACTGAAATCTGAACGCCTAACTCGTTATCAAGGTGTTAATTTGTATGTTAAGAACTTGGATGATACAATTGATGATGAACGTCTGCGCAAAGAATTTGCACCATTTGGGACTATTACGTCTGCGAAGGTAATGCTAGAAGATGGACGTAGTAAAGGCTTTGGGTTTGTTTGTTTTTCTTCTCCTGAAGAAGCTACCAAAGCAGTTACTGAGATGAATGGACGTATTGTAGGAACTAAACCATTGTATGTTGCACTAGCTCAACGCAAGGAGGACCGTAAAGCGCACTTAACATCTCAGTACATGCAGAGAATGGCTAGTATGCGCATGCAGCAAATGGGTCAAATATTCCAGCCTAGTGGTGCAAGTGGATTTTTTGTACCATCGCTCCCTCCAGCACCACGTTACTATGGACCAGCTCAAATGACTCAAATAAGACCTACAACTCCAAGGTGGAATGCACAGCCCCCTGTAAGGCCTAGCACACAAGCAGCATCTGCATATGCAAATATGCAACCCACTTACAGACCAGCACCGAGAGCTCCGGCCCAATCTACAATAAGAACATCGTTAGGAGCAAGACCCATAACTGGACAGCAAGGAATTGCTGCTGCTGCTGCATCTATTCGTCCACCTTTAGTATCTCAAAGCAGTCGCCCAGCCAATTATAAATATACACCAAACATGCGCAACCCACCAGCTCCACAACCAGCAGTGCATATCCAAGGACAAGAACCTCTGACCTCTACTATGCTTGCAGCAGCACCACTCCAGGAACAGAAACAGATGCTTGGGGAACGACTGTTTCCACTTATTCAGAGAATGCACCCTGATTTGGCTGGGAAAATAACTGGTATGCTTTTGGAAATTGATAATTCAGAACTTCTTCATATGTTAGAACATGCAGAGTCTCTGAAAGCTAAAGTAGATGAAGCTGTTGCTGTACTGCAGGCTCATCAAGCTAAACAACAAGCAACTAAAAAAGAATAA
Protein
MNPGPPSYPMASLYVGDLHSDITEAMLFEKFSTAGPVLSIRVCRDMITRRSLGYAYVNFQQPTDAERALDTMNFDIIKGRPIRIMWSQRDPSLRKSGVGNVFIKNLDKSIDNKALFDTFSAFGNILSCKVAQDETGASKGYGFVHFETEEAANKSIEKVNGMLLNGKMVYVGRFIPRKEREKELGEKAKLFANVYVKNFGEDFSDEMLKDMFEKYGRITSHKVMYKDDGNSRGFGFVAFEDPDAAERASIELNGKELVEGKPLYVGRAQKKAERQKELKRKFEQLKSERLTRYQGVNLYVKNLDDTIDDERLRKEFAPFGTITSAKVMLEDGRSKGFGFVCFSSPEEATKAVTEMNGRIVGTKPLYVALAQRKEDRKAHLTSQYMQRMASMRMQQMGQIFQPSGASGFFVPSLPPAPRYYGPAQMTQIRPTTPRWNAQPPVRPSTQAASAYANMQPTYRPAPRAPAQSTIRTSLGARPITGQQGIAAAAASIRPPLVSQSSRPANYKYTPNMRNPPAPQPAVHIQGQEPLTSTMLAAAPLQEQKQMLGERLFPLIQRMHPDLAGKITGMLLEIDNSELLHMLEHAESLKAKVDEAVAVLQAHQAKQQATKKE
Summary
Description
Binds the poly(A) tail of mRNA.
Similarity
Belongs to the polyadenylate-binding protein type-1 family.
Uniprot
Q0N2S4
A0A2W1B7S2
A0A2A4JNP3
A0A2H1W6T4
I4DLZ9
I4DII6
+ More
A0A194RH12 A0A194PQ29 S4NY68 A0A3G1T181 A0A212FIZ3 A0A0L7KP79 A0A2J7QJ01 A0A2J7QKS7 A0A067REV7 A0A2A3E2F3 V9IIU3 A0A158P240 A0A2A4JM96 A0A1W4XB05 A0A154NZZ8 A0A1W4X096 A0A151IEQ6 A0A1W4X9L5 K7J4C2 A0A1Y1LJP2 A0A232F8M2 A0A139WP55 A0A026VW79 A0A1W4XAT7 A0A1B6C293 E2B282 K7J4C3 K7J4C1 A0A195AW07 A0A0A9XTF5 A0A195AVN5 A0A195FJ12 A0A158P241 A0A151IEM4 A0A195DMD4 F4W6C1 A0A151XCR0 A0A0A9XQF4 A0A195FK67 D6W7F7 F4W6C2 V5G5Q5 E0W4B6 J3JVB3 A0A1Y1NB17 A0A088ASD5 U5EVS1 A0A1D1XE96 A0A1L8DHH2 A0A1L8DHB1 A0A1B0CLG4 A0A336K6T9 T1E7C5 A0A182FTK5 A0A2M3YXC2 A0A2M3ZY95 A0A2M4BH36 W5JFF8 A0A2M4BGV2 Q1HR66 A0A1L8DH66 A0A023EVL3 H9J1T2 A0A182IM26 X1WI92 B0WMK0 A0A1Q3FCL3 A0A182Q873 A0A182NSX0 A0A1B6IGJ5 A0A2H8TQ52 A0A084WLT3 A0A1W7R9X3 A0A1J1HNU4 A0A182RLS8 A0A182YAV4 A0A182WUC9 A0A182VMS8 A0A1S4H5Q0 A0A2C9GRC7 A0A087T5F4 A0A2I9LPG2 A0A0P4WRL4 A0A385L4C5 A0A2L2Y0U4 A0A182P5C9 Q7QH99 A0A0N8ER37 A0A0K8TQ12 A0A0P6HXB6 A0A0N8CG33 A0A0P5ZHV1 J9KAC6 A0A1B6EQV5
A0A194RH12 A0A194PQ29 S4NY68 A0A3G1T181 A0A212FIZ3 A0A0L7KP79 A0A2J7QJ01 A0A2J7QKS7 A0A067REV7 A0A2A3E2F3 V9IIU3 A0A158P240 A0A2A4JM96 A0A1W4XB05 A0A154NZZ8 A0A1W4X096 A0A151IEQ6 A0A1W4X9L5 K7J4C2 A0A1Y1LJP2 A0A232F8M2 A0A139WP55 A0A026VW79 A0A1W4XAT7 A0A1B6C293 E2B282 K7J4C3 K7J4C1 A0A195AW07 A0A0A9XTF5 A0A195AVN5 A0A195FJ12 A0A158P241 A0A151IEM4 A0A195DMD4 F4W6C1 A0A151XCR0 A0A0A9XQF4 A0A195FK67 D6W7F7 F4W6C2 V5G5Q5 E0W4B6 J3JVB3 A0A1Y1NB17 A0A088ASD5 U5EVS1 A0A1D1XE96 A0A1L8DHH2 A0A1L8DHB1 A0A1B0CLG4 A0A336K6T9 T1E7C5 A0A182FTK5 A0A2M3YXC2 A0A2M3ZY95 A0A2M4BH36 W5JFF8 A0A2M4BGV2 Q1HR66 A0A1L8DH66 A0A023EVL3 H9J1T2 A0A182IM26 X1WI92 B0WMK0 A0A1Q3FCL3 A0A182Q873 A0A182NSX0 A0A1B6IGJ5 A0A2H8TQ52 A0A084WLT3 A0A1W7R9X3 A0A1J1HNU4 A0A182RLS8 A0A182YAV4 A0A182WUC9 A0A182VMS8 A0A1S4H5Q0 A0A2C9GRC7 A0A087T5F4 A0A2I9LPG2 A0A0P4WRL4 A0A385L4C5 A0A2L2Y0U4 A0A182P5C9 Q7QH99 A0A0N8ER37 A0A0K8TQ12 A0A0P6HXB6 A0A0N8CG33 A0A0P5ZHV1 J9KAC6 A0A1B6EQV5
Pubmed
28756777
22651552
26354079
23622113
22118469
26227816
+ More
24845553 21347285 20075255 28004739 28648823 18362917 19820115 24508170 30249741 20798317 25401762 26823975 21719571 20566863 22516182 23537049 20920257 23761445 17204158 17510324 24945155 19121390 24438588 25244985 12364791 29248469 26561354 26369729
24845553 21347285 20075255 28004739 28648823 18362917 19820115 24508170 30249741 20798317 25401762 26823975 21719571 20566863 22516182 23537049 20920257 23761445 17204158 17510324 24945155 19121390 24438588 25244985 12364791 29248469 26561354 26369729
EMBL
DQ646407
ABH10797.1
KZ150495
PZC70675.1
NWSH01000990
PCG73204.1
+ More
ODYU01006578 SOQ48542.1 AK402317 BAM18939.1 AK401104 BAM17726.1 KQ460207 KPJ16729.1 KQ459597 KPI94844.1 GAIX01013920 JAA78640.1 MG846895 AXY94747.1 AGBW02008321 OWR53699.1 JTDY01007562 KOB65113.1 NEVH01013563 PNF28560.1 NEVH01013256 PNF29191.1 KK852549 KDR21568.1 KZ288425 PBC25923.1 JR046784 AEY60249.1 ADTU01006976 PCG73205.1 KQ434786 KZC05162.1 KQ977858 KYM99295.1 AAZX01006389 AAZX01006683 GEZM01054088 JAV73853.1 NNAY01000718 OXU26809.1 KQ971307 KYB29724.1 KK107790 QOIP01000010 EZA47761.1 RLU18011.1 GEDC01029963 JAS07335.1 GL445067 EFN60208.1 KQ976731 KYM76242.1 GBHO01020638 GBRD01010657 GDHC01004198 JAG22966.1 JAG55167.1 JAQ14431.1 KYM76241.1 KQ981523 KYN40393.1 KYM99294.1 KQ980724 KYN14060.1 GL887707 EGI70256.1 KQ982297 KYQ58171.1 GBHO01020637 GBRD01001909 GDHC01000230 JAG22967.1 JAG63912.1 JAQ18399.1 KYN40394.1 EFA11220.1 EGI70257.1 GALX01003056 JAB65410.1 DS235886 EEB20472.1 APGK01047223 BT127180 KB741077 KB631792 AEE62142.1 ENN74236.1 ERL86135.1 GEZM01010785 JAV93835.1 GANO01000888 JAB58983.1 GDJX01027204 JAT40732.1 GFDF01008284 JAV05800.1 GFDF01008283 JAV05801.1 AJWK01017220 AJWK01017221 UFQS01000128 UFQT01000128 SSX00191.1 SSX20571.1 GAMD01003167 JAA98423.1 GGFM01000161 MBW20912.1 GGFK01000203 MBW33524.1 GGFJ01003120 MBW52261.1 ADMH02001295 ETN63107.1 GGFJ01003121 MBW52262.1 DQ440228 CH477655 ABF18261.1 EAT37709.1 GFDF01008282 JAV05802.1 GAPW01000522 GAPW01000521 JAC13076.1 BABH01008005 BABH01008006 ABLF02016904 DS231998 EDS31071.1 GFDL01009817 JAV25228.1 AXCN02002137 GECU01021667 JAS86039.1 GFXV01004519 MBW16324.1 ATLV01024277 KE525351 KFB51177.1 GFAH01000442 JAV47947.1 CVRI01000011 CRK89202.1 AAAB01008816 APCN01001462 KK113500 KFM60343.1 GFWZ01000291 MBW20281.1 GDRN01041197 JAI67105.1 MH538277 AYA22653.1 IAAA01006564 LAA01816.1 EAA05186.3 GDIQ01002172 JAN92565.1 GDAI01001380 JAI16223.1 GDIQ01028451 JAN66286.1 GDIP01135183 JAL68531.1 GDIP01043332 JAM60383.1 ABLF02010624 ABLF02010625 GECZ01029424 JAS40345.1
ODYU01006578 SOQ48542.1 AK402317 BAM18939.1 AK401104 BAM17726.1 KQ460207 KPJ16729.1 KQ459597 KPI94844.1 GAIX01013920 JAA78640.1 MG846895 AXY94747.1 AGBW02008321 OWR53699.1 JTDY01007562 KOB65113.1 NEVH01013563 PNF28560.1 NEVH01013256 PNF29191.1 KK852549 KDR21568.1 KZ288425 PBC25923.1 JR046784 AEY60249.1 ADTU01006976 PCG73205.1 KQ434786 KZC05162.1 KQ977858 KYM99295.1 AAZX01006389 AAZX01006683 GEZM01054088 JAV73853.1 NNAY01000718 OXU26809.1 KQ971307 KYB29724.1 KK107790 QOIP01000010 EZA47761.1 RLU18011.1 GEDC01029963 JAS07335.1 GL445067 EFN60208.1 KQ976731 KYM76242.1 GBHO01020638 GBRD01010657 GDHC01004198 JAG22966.1 JAG55167.1 JAQ14431.1 KYM76241.1 KQ981523 KYN40393.1 KYM99294.1 KQ980724 KYN14060.1 GL887707 EGI70256.1 KQ982297 KYQ58171.1 GBHO01020637 GBRD01001909 GDHC01000230 JAG22967.1 JAG63912.1 JAQ18399.1 KYN40394.1 EFA11220.1 EGI70257.1 GALX01003056 JAB65410.1 DS235886 EEB20472.1 APGK01047223 BT127180 KB741077 KB631792 AEE62142.1 ENN74236.1 ERL86135.1 GEZM01010785 JAV93835.1 GANO01000888 JAB58983.1 GDJX01027204 JAT40732.1 GFDF01008284 JAV05800.1 GFDF01008283 JAV05801.1 AJWK01017220 AJWK01017221 UFQS01000128 UFQT01000128 SSX00191.1 SSX20571.1 GAMD01003167 JAA98423.1 GGFM01000161 MBW20912.1 GGFK01000203 MBW33524.1 GGFJ01003120 MBW52261.1 ADMH02001295 ETN63107.1 GGFJ01003121 MBW52262.1 DQ440228 CH477655 ABF18261.1 EAT37709.1 GFDF01008282 JAV05802.1 GAPW01000522 GAPW01000521 JAC13076.1 BABH01008005 BABH01008006 ABLF02016904 DS231998 EDS31071.1 GFDL01009817 JAV25228.1 AXCN02002137 GECU01021667 JAS86039.1 GFXV01004519 MBW16324.1 ATLV01024277 KE525351 KFB51177.1 GFAH01000442 JAV47947.1 CVRI01000011 CRK89202.1 AAAB01008816 APCN01001462 KK113500 KFM60343.1 GFWZ01000291 MBW20281.1 GDRN01041197 JAI67105.1 MH538277 AYA22653.1 IAAA01006564 LAA01816.1 EAA05186.3 GDIQ01002172 JAN92565.1 GDAI01001380 JAI16223.1 GDIQ01028451 JAN66286.1 GDIP01135183 JAL68531.1 GDIP01043332 JAM60383.1 ABLF02010624 ABLF02010625 GECZ01029424 JAS40345.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
UP000235965
+ More
UP000027135 UP000242457 UP000005205 UP000192223 UP000076502 UP000078542 UP000002358 UP000215335 UP000007266 UP000053097 UP000279307 UP000000311 UP000078540 UP000078541 UP000078492 UP000007755 UP000075809 UP000009046 UP000019118 UP000030742 UP000005203 UP000092461 UP000069272 UP000000673 UP000008820 UP000005204 UP000075880 UP000007819 UP000002320 UP000075886 UP000075884 UP000030765 UP000183832 UP000075900 UP000076408 UP000076407 UP000075903 UP000075840 UP000054359 UP000075885 UP000007062
UP000027135 UP000242457 UP000005205 UP000192223 UP000076502 UP000078542 UP000002358 UP000215335 UP000007266 UP000053097 UP000279307 UP000000311 UP000078540 UP000078541 UP000078492 UP000007755 UP000075809 UP000009046 UP000019118 UP000030742 UP000005203 UP000092461 UP000069272 UP000000673 UP000008820 UP000005204 UP000075880 UP000007819 UP000002320 UP000075886 UP000075884 UP000030765 UP000183832 UP000075900 UP000076408 UP000076407 UP000075903 UP000075840 UP000054359 UP000075885 UP000007062
Interpro
Gene 3D
ProteinModelPortal
Q0N2S4
A0A2W1B7S2
A0A2A4JNP3
A0A2H1W6T4
I4DLZ9
I4DII6
+ More
A0A194RH12 A0A194PQ29 S4NY68 A0A3G1T181 A0A212FIZ3 A0A0L7KP79 A0A2J7QJ01 A0A2J7QKS7 A0A067REV7 A0A2A3E2F3 V9IIU3 A0A158P240 A0A2A4JM96 A0A1W4XB05 A0A154NZZ8 A0A1W4X096 A0A151IEQ6 A0A1W4X9L5 K7J4C2 A0A1Y1LJP2 A0A232F8M2 A0A139WP55 A0A026VW79 A0A1W4XAT7 A0A1B6C293 E2B282 K7J4C3 K7J4C1 A0A195AW07 A0A0A9XTF5 A0A195AVN5 A0A195FJ12 A0A158P241 A0A151IEM4 A0A195DMD4 F4W6C1 A0A151XCR0 A0A0A9XQF4 A0A195FK67 D6W7F7 F4W6C2 V5G5Q5 E0W4B6 J3JVB3 A0A1Y1NB17 A0A088ASD5 U5EVS1 A0A1D1XE96 A0A1L8DHH2 A0A1L8DHB1 A0A1B0CLG4 A0A336K6T9 T1E7C5 A0A182FTK5 A0A2M3YXC2 A0A2M3ZY95 A0A2M4BH36 W5JFF8 A0A2M4BGV2 Q1HR66 A0A1L8DH66 A0A023EVL3 H9J1T2 A0A182IM26 X1WI92 B0WMK0 A0A1Q3FCL3 A0A182Q873 A0A182NSX0 A0A1B6IGJ5 A0A2H8TQ52 A0A084WLT3 A0A1W7R9X3 A0A1J1HNU4 A0A182RLS8 A0A182YAV4 A0A182WUC9 A0A182VMS8 A0A1S4H5Q0 A0A2C9GRC7 A0A087T5F4 A0A2I9LPG2 A0A0P4WRL4 A0A385L4C5 A0A2L2Y0U4 A0A182P5C9 Q7QH99 A0A0N8ER37 A0A0K8TQ12 A0A0P6HXB6 A0A0N8CG33 A0A0P5ZHV1 J9KAC6 A0A1B6EQV5
A0A194RH12 A0A194PQ29 S4NY68 A0A3G1T181 A0A212FIZ3 A0A0L7KP79 A0A2J7QJ01 A0A2J7QKS7 A0A067REV7 A0A2A3E2F3 V9IIU3 A0A158P240 A0A2A4JM96 A0A1W4XB05 A0A154NZZ8 A0A1W4X096 A0A151IEQ6 A0A1W4X9L5 K7J4C2 A0A1Y1LJP2 A0A232F8M2 A0A139WP55 A0A026VW79 A0A1W4XAT7 A0A1B6C293 E2B282 K7J4C3 K7J4C1 A0A195AW07 A0A0A9XTF5 A0A195AVN5 A0A195FJ12 A0A158P241 A0A151IEM4 A0A195DMD4 F4W6C1 A0A151XCR0 A0A0A9XQF4 A0A195FK67 D6W7F7 F4W6C2 V5G5Q5 E0W4B6 J3JVB3 A0A1Y1NB17 A0A088ASD5 U5EVS1 A0A1D1XE96 A0A1L8DHH2 A0A1L8DHB1 A0A1B0CLG4 A0A336K6T9 T1E7C5 A0A182FTK5 A0A2M3YXC2 A0A2M3ZY95 A0A2M4BH36 W5JFF8 A0A2M4BGV2 Q1HR66 A0A1L8DH66 A0A023EVL3 H9J1T2 A0A182IM26 X1WI92 B0WMK0 A0A1Q3FCL3 A0A182Q873 A0A182NSX0 A0A1B6IGJ5 A0A2H8TQ52 A0A084WLT3 A0A1W7R9X3 A0A1J1HNU4 A0A182RLS8 A0A182YAV4 A0A182WUC9 A0A182VMS8 A0A1S4H5Q0 A0A2C9GRC7 A0A087T5F4 A0A2I9LPG2 A0A0P4WRL4 A0A385L4C5 A0A2L2Y0U4 A0A182P5C9 Q7QH99 A0A0N8ER37 A0A0K8TQ12 A0A0P6HXB6 A0A0N8CG33 A0A0P5ZHV1 J9KAC6 A0A1B6EQV5
PDB
6R5K
E-value=5.75103e-115,
Score=1061
Ontologies
PATHWAY
GO
Topology
Subcellular location
Cytoplasm
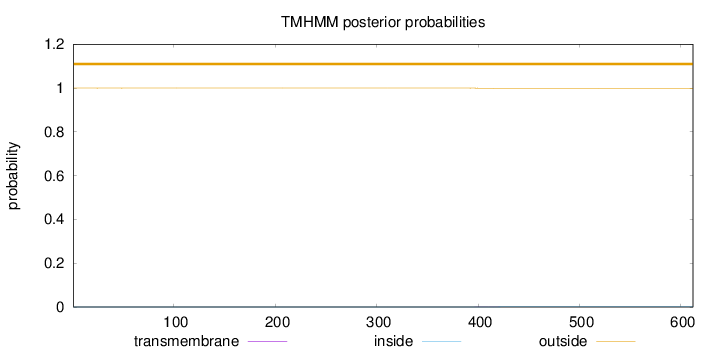
Length:
612
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01208
Exp number, first 60 AAs:
0.0006
Total prob of N-in:
0.00014
outside
1 - 612
Population Genetic Test Statistics
Pi
332.89918
Theta
154.2202
Tajima's D
3.827825
CLR
0.141371
CSRT
0.9978001099945
Interpretation
Uncertain
