Gene
KWMTBOMO15927
Pre Gene Modal
BGIBMGA010315
Annotation
Gag-like_protein_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 2.416
Sequence
CDS
ATGGGGGTCACTACCGAGGACCCTGGTGGCGAGAGCTGTGCCTCGCTAGGGAGCTCAAAGGCGGAGATCAATGCCGCCCGAAGAGAGCAGCGCAAAGCTGTCGCGGCCGACGAAGTGTCGGAAATGGCCCGACGCGCTCGCGAGCGACGCGCTACCCTAGCGGCGGAAGGGGGGGAACCATCTGCGGTGGCCCTAAGTCAGCTTGCCTTGGACGGGGTGGACTTAATCCTGAAGGTTGCCACCAAATCCGGCAGCCTTAAGGGCACGTTTACCCGCGGTCTGAAAGAAGCTGCCGCGGACATTAAGGAGGCGATTGGCATCCTCCTCAACAGGACGGCCTCTGACGAGGTTGCGAAGTTGCAGGAGGAAAACAGCCGTCTCCGAAATGATATGGAGGACCTCCGGCGACGAGTCACTGCGTTGAGCGAGCAGCAGCAGCGGCGTACGTCCACTGATGCCGCATCGGTAGTGGCCCCGGCTCCCGCACCGAGACCGACAAGCACTCATACCGACGACGAGGTCGAGCGCATTGTCCGGCTCTGTATGCTCCAGTGCGGGAGCATGGTCAATGCTCGTATGGAGGCAATTTCTCGGCGCCTCCCTGCGGAAATCCTCCGTCCGCCACTAGCGGCCGATACACGGCGGAGGGCTGAGGAGCCGCCGAGACCTAAGCCTAGGGAGGGGAAGCCCGCGGAGGGTGTTAAAAAGCCCGTCGAGGGAGCCCCATCGAGCGATCAGCCCACGACTGCTGGGTCTAAGGGTGAGACCTGGGTTACAGTCGTGGGCCGCAAGAAGGCACGCAGGGTCGCCAAGGCGGCCTCAGCAGCAACGCATGCCCCCGGCCAAACCGCAAAGGCGGTTGCGCAGCCTGCGCGGCGGGCTGCCAAAGGCGGTCGCAAAGGACCGGCGATACGTGCTCCGCGTTCCGAAGCTGTGACGCTCACGCTACAGCCTGGAGCTGCGGAGCGCGGCGTAACGTACCAGTCGGTCATCGCCGAAGCAAAGGCCAAGATCAAATTATCAGATCTTGGTCTTCAGGCCGTCACCCACAGGCAGGCTGCCACGGGTGCACGGTTGTTCGAGGTGGCTGGTACTACGAGTGGCAGTGCCGAAAAAGCGGACGCTCTGGCCGCCAAGATGAGGGAGGTCCTCAGCCCCGAGGACGTCCGGGTCTCCAGGCCAGTGAAAACCGCAGAGGTGCGGATTACTGGCCTGGATGACTCCGTGACCTCTGAGGAGGTGGTAGCGGCCGTTGCCCGAAGTGGAGAGTGTCCATCGGACAAGGTGCGGGCCGGCGATATACGCACCGACGCCACCGGACTCGGCGTGGCCTGGGTTCGGTGCCCTGTAGCGTCGGCGAAAAAGATCGCCTTAAGCGGCAGATTGCTGCTCTTGAAGCCGAGAATCACATCTATCACCTGCGTTATAAATGCGTCGCGGCCCGTATCGTGTCACATTCGTTAA
Protein
MGVTTEDPGGESCASLGSSKAEINAARREQRKAVAADEVSEMARRARERRATLAAEGGEPSAVALSQLALDGVDLILKVATKSGSLKGTFTRGLKEAAADIKEAIGILLNRTASDEVAKLQEENSRLRNDMEDLRRRVTALSEQQQRRTSTDAASVVAPAPAPRPTSTHTDDEVERIVRLCMLQCGSMVNARMEAISRRLPAEILRPPLAADTRRRAEEPPRPKPREGKPAEGVKKPVEGAPSSDQPTTAGSKGETWVTVVGRKKARRVAKAASAATHAPGQTAKAVAQPARRAAKGGRKGPAIRAPRSEAVTLTLQPGAAERGVTYQSVIAEAKAKIKLSDLGLQAVTHRQAATGARLFEVAGTTSGSAEKADALAAKMREVLSPEDVRVSRPVKTAEVRITGLDDSVTSEEVVAAVARSGECPSDKVRAGDIRTDATGLGVAWVRCPVASAKKIALSGRLLLLKPRITSITCVINASRPVSCHIR
Summary
Uniprot
Pubmed
EMBL
Proteomes
Pfam
PF00098 zf-CCHC
SUPFAM
SSF57756
SSF57756
ProteinModelPortal
Ontologies
KEGG
Topology
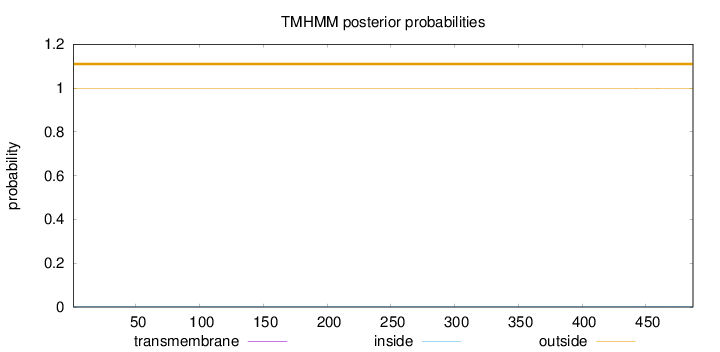
Length:
487
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01071
Exp number, first 60 AAs:
0.00046
Total prob of N-in:
0.00124
outside
1 - 487
Population Genetic Test Statistics
Pi
249.947843
Theta
144.443504
Tajima's D
2.280452
CLR
188.096321
CSRT
0.921753912304385
Interpretation
Uncertain
