Gene
KWMTBOMO15920 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001320
Annotation
PREDICTED:_trypsin?_alkaline_B-like_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 1.992 PlasmaMembrane Reliability : 2.449
Sequence
CDS
ATGGCTTCTAAGGTGTTGTACTTGATGCTCGCATTTGTGGCCGCCGTATCGGCAGCGCCCAACACTCGTCTCGTGGGCGGTGAGCACACCACCATAGAGCAGTTTCCGTACATCGTCGCCCTGGTGTACTATTACCCCAGGCCTGCCATCAGAGTACAAAGATGCGTAGGTTCGTTGATCTCATCTTGGCACGTAGTGACTTCAGGCTATTGTTTCACTGGCGCTATCCTTGATAATATGGAGATCCGAGCTGGCTCTACCAACGCCTTATCCGGTGGTACCGTGGTCCGTGTAGGACAGGTGACCAAGTACCCTGGGTACGTTGAAGAGCCAAAGTCTGGTGATCTTGCCGTGATCCGCTTGGCTACGGCCCTGGGCATCAGCAACACGGTCAACGTGCTCTTCCTTCCTCCTCAAAACACTTTTATCCCCGATGGCACTTCTCTTACCACCGTTAGCTGGGGTTTTGAATACGTAGGTGGCCCACAGCTGGAGACCCTGAAAACCATCACCCAAAGAAAGATCAACATTGAGCGATGCAAAGCCATTTACAGTGGTTCTACTAAAGTCGACATTTCTGACCGCGTGATCTGCGCCAACGAGGTCGGCCGCAGCACTTGCTTCGGCGACTCCGGAGCGCCCATGGTCATCGGCCAAGTTCTAGTCGGCACTGCTTCCTACCACGAGGGCTGCGGTGACGACGACCACCCCGATGTCTTCACCAGAATCGACACCTATACCAACTGGATCATGCAGGTGGCTACCGCGCCTGGAAGGAGTGGCTCCGAAGCGAAGATAGCACCTGTACAATTTTAA
Protein
MASKVLYLMLAFVAAVSAAPNTRLVGGEHTTIEQFPYIVALVYYYPRPAIRVQRCVGSLISSWHVVTSGYCFTGAILDNMEIRAGSTNALSGGTVVRVGQVTKYPGYVEEPKSGDLAVIRLATALGISNTVNVLFLPPQNTFIPDGTSLTTVSWGFEYVGGPQLETLKTITQRKINIERCKAIYSGSTKVDISDRVICANEVGRSTCFGDSGAPMVIGQVLVGTASYHEGCGDDDHPDVFTRIDTYTNWIMQVATAPGRSGSEAKIAPVQF
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9IVN9
A0A194Q9W6
A0A1E1W615
A0A2H1WPC3
A0A2A4JX60
D2Y4U5
+ More
A0A023JNS6 B6CME7 A0A0N1IE76 I6SW41 C9W8I7 A0A2Z5U7J9 I6TRU2 A0A0L7LSM8 B6D1Q8 A0A089RSX5 I6TRT9 Q4L1L0 A0A0N1I8C7 Q9Y7A9 E9FBE3 A0A2W1BJP2 A0A2A4JTP3 A0A0L7KYH0 A0A194QET8 J7IM02 A0A194PP33 A0A0B4EQ31 B4X996 B4X9A2 B1NLD8 A0A2A4JTX8 A0A2H1WEE4 J7IN23 A0A194PVB6 A0A0N1PHJ0 A0A0M4M544 A0A0L7L163 A0A1S4H7M9 A0A2W1BE44 A0A182UPH4 Q6R560 C9W8J1 A0A2W1BTM0 A0A2W1BJK7 A0A212FL72 B4X995 O18435 A0A182LYQ9 B0ZBM9 A0A340TBF8 I7D523 J7IJ83 A0A182LPG3 A0A2H1WTV1 A0A194QDC5 A0A0B4ERM0 A0A2A4K5G7 C9W8J5 A0A0D9NL96 A0A194QJ28 C9W8I2 A0A2A4JCA8 A0A2A4K648 A0A182M5F2 A0A182KHH7 A0A2H1VGQ8 A0A395HH54 I6TMW8 A0A182MZ05 A0A182UGL3 A0A2H3FQP9 Q9NB91 Q4L1K1 Q7Q344 D3TKS0 A0A2A4JRB9 D2A2R7 A0A2H1VGP3 B6CME9 A0A182T760 Q4L1K4 C9W8F7 O18447 C9W8I6 A0A182T724 A0A194PP38 B0WBW0 J7IFG7 J3JW59 A0A2A4K636 N6TW63 U4TXR1 V9LL27 B4X9A0 A0A182YPE1 A0A2A4K6E9
A0A023JNS6 B6CME7 A0A0N1IE76 I6SW41 C9W8I7 A0A2Z5U7J9 I6TRU2 A0A0L7LSM8 B6D1Q8 A0A089RSX5 I6TRT9 Q4L1L0 A0A0N1I8C7 Q9Y7A9 E9FBE3 A0A2W1BJP2 A0A2A4JTP3 A0A0L7KYH0 A0A194QET8 J7IM02 A0A194PP33 A0A0B4EQ31 B4X996 B4X9A2 B1NLD8 A0A2A4JTX8 A0A2H1WEE4 J7IN23 A0A194PVB6 A0A0N1PHJ0 A0A0M4M544 A0A0L7L163 A0A1S4H7M9 A0A2W1BE44 A0A182UPH4 Q6R560 C9W8J1 A0A2W1BTM0 A0A2W1BJK7 A0A212FL72 B4X995 O18435 A0A182LYQ9 B0ZBM9 A0A340TBF8 I7D523 J7IJ83 A0A182LPG3 A0A2H1WTV1 A0A194QDC5 A0A0B4ERM0 A0A2A4K5G7 C9W8J5 A0A0D9NL96 A0A194QJ28 C9W8I2 A0A2A4JCA8 A0A2A4K648 A0A182M5F2 A0A182KHH7 A0A2H1VGQ8 A0A395HH54 I6TMW8 A0A182MZ05 A0A182UGL3 A0A2H3FQP9 Q9NB91 Q4L1K1 Q7Q344 D3TKS0 A0A2A4JRB9 D2A2R7 A0A2H1VGP3 B6CME9 A0A182T760 Q4L1K4 C9W8F7 O18447 C9W8I6 A0A182T724 A0A194PP38 B0WBW0 J7IFG7 J3JW59 A0A2A4K636 N6TW63 U4TXR1 V9LL27 B4X9A0 A0A182YPE1 A0A2A4K6E9
Pubmed
EMBL
BABH01039650
KQ459249
KPJ02313.1
GDQN01008638
JAT82416.1
ODYU01010075
+ More
SOQ54919.1 NWSH01000442 PCG76399.1 GU323797 ADA83702.1 KF791044 AHL46496.1 EU325547 ACB54937.1 LADJ01028292 KPJ21177.1 JQ904130 AFM77760.1 FJ205434 ACR16000.1 AP017543 BBB06944.1 JQ904131 AFM77761.1 JTDY01000226 KOB78196.1 EU673451 ACI45414.1 KM083776 AIR09757.1 JQ904126 AFM77756.1 AY587155 AAT95347.1 KQ460409 KPJ14944.1 AF130865 AAD29675.1 ADNJ02000009 EFY94881.1 KZ150010 PZC75088.1 NWSH01000631 PCG75146.1 JTDY01004416 KOB68185.1 KQ459167 KPJ03460.1 JX195650 AFQ37933.1 KQ459597 KPI95067.1 AZNG01000025 KID64135.1 EF531632 ABR88243.1 EF531638 ABR88249.1 EF600053 ABU98618.1 PZC75086.1 PCG75148.1 ODYU01007983 SOQ51222.1 JX195648 AFQ37931.1 KPI95070.1 KQ460226 KPJ16473.1 KR024672 ALE15214.1 JTDY01003642 KOB69233.1 AAAB01008966 KZ150104 PZC73442.1 AY513650 AAR98919.1 FJ205440 ACR16004.1 PZC75083.1 PZC73447.1 AGBW02007905 OWR54439.1 EF531631 EF531633 ABR88242.1 ABR88244.1 Y12270 CAA72949.1 AXCM01002130 EU339295 ABZ04009.1 JN793545 AFO68325.1 JX195651 AFQ37934.1 ODYU01011017 SOQ56495.1 KPJ03462.1 AZNF01000008 KID64620.1 NWSH01000106 PCG79485.1 FJ205445 ACR16008.2 KE384755 KJK74714.1 KPJ03461.1 FJ205428 ACR15995.1 NWSH01002022 PCG69396.1 PCG79486.1 AXCM01007963 AXCM01007964 ODYU01002467 SOQ39996.1 KZ824351 RAL06823.1 JQ904123 AFM77753.1 MVNX01000034 PBP24814.1 AF261971 AF261990 AAF74733.1 AY587164 AAT95356.1 EAA13086.4 EZ422007 ADD18430.1 NWSH01000830 PCG73960.1 KQ971339 EFA02919.1 SOQ39998.1 EU325549 ACB54939.1 AY587161 AAT95353.1 FJ205402 ACR15970.1 HM209426 Y12283 ADI32887.1 CAA72962.1 PZC73444.1 FJ205433 ACR15999.2 KPI95072.1 DS231882 EDS42890.1 JX195652 AFQ37935.1 BT127477 AEE62439.1 PCG79476.1 APGK01052535 KB741213 ENN72611.1 KB631625 ERL84793.1 JQ340915 AFP74111.1 EF531636 ABR88247.1 PCG79483.1 PCG79488.1
SOQ54919.1 NWSH01000442 PCG76399.1 GU323797 ADA83702.1 KF791044 AHL46496.1 EU325547 ACB54937.1 LADJ01028292 KPJ21177.1 JQ904130 AFM77760.1 FJ205434 ACR16000.1 AP017543 BBB06944.1 JQ904131 AFM77761.1 JTDY01000226 KOB78196.1 EU673451 ACI45414.1 KM083776 AIR09757.1 JQ904126 AFM77756.1 AY587155 AAT95347.1 KQ460409 KPJ14944.1 AF130865 AAD29675.1 ADNJ02000009 EFY94881.1 KZ150010 PZC75088.1 NWSH01000631 PCG75146.1 JTDY01004416 KOB68185.1 KQ459167 KPJ03460.1 JX195650 AFQ37933.1 KQ459597 KPI95067.1 AZNG01000025 KID64135.1 EF531632 ABR88243.1 EF531638 ABR88249.1 EF600053 ABU98618.1 PZC75086.1 PCG75148.1 ODYU01007983 SOQ51222.1 JX195648 AFQ37931.1 KPI95070.1 KQ460226 KPJ16473.1 KR024672 ALE15214.1 JTDY01003642 KOB69233.1 AAAB01008966 KZ150104 PZC73442.1 AY513650 AAR98919.1 FJ205440 ACR16004.1 PZC75083.1 PZC73447.1 AGBW02007905 OWR54439.1 EF531631 EF531633 ABR88242.1 ABR88244.1 Y12270 CAA72949.1 AXCM01002130 EU339295 ABZ04009.1 JN793545 AFO68325.1 JX195651 AFQ37934.1 ODYU01011017 SOQ56495.1 KPJ03462.1 AZNF01000008 KID64620.1 NWSH01000106 PCG79485.1 FJ205445 ACR16008.2 KE384755 KJK74714.1 KPJ03461.1 FJ205428 ACR15995.1 NWSH01002022 PCG69396.1 PCG79486.1 AXCM01007963 AXCM01007964 ODYU01002467 SOQ39996.1 KZ824351 RAL06823.1 JQ904123 AFM77753.1 MVNX01000034 PBP24814.1 AF261971 AF261990 AAF74733.1 AY587164 AAT95356.1 EAA13086.4 EZ422007 ADD18430.1 NWSH01000830 PCG73960.1 KQ971339 EFA02919.1 SOQ39998.1 EU325549 ACB54939.1 AY587161 AAT95353.1 FJ205402 ACR15970.1 HM209426 Y12283 ADI32887.1 CAA72962.1 PZC73444.1 FJ205433 ACR15999.2 KPI95072.1 DS231882 EDS42890.1 JX195652 AFQ37935.1 BT127477 AEE62439.1 PCG79476.1 APGK01052535 KB741213 ENN72611.1 KB631625 ERL84793.1 JQ340915 AFP74111.1 EF531636 ABR88247.1 PCG79483.1 PCG79488.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000037510
UP000002498
+ More
UP000031181 UP000075903 UP000007151 UP000075883 UP000076407 UP000075882 UP000031186 UP000054544 UP000075881 UP000248961 UP000075884 UP000075902 UP000218527 UP000007062 UP000007266 UP000075901 UP000002320 UP000019118 UP000030742 UP000076408
UP000031181 UP000075903 UP000007151 UP000075883 UP000076407 UP000075882 UP000031186 UP000054544 UP000075881 UP000248961 UP000075884 UP000075902 UP000218527 UP000007062 UP000007266 UP000075901 UP000002320 UP000019118 UP000030742 UP000076408
PRIDE
Interpro
IPR001254
Trypsin_dom
+ More
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR018114 TRYPSIN_HIS
IPR033116 TRYPSIN_SER
IPR012224 Pept_S1A_FX
IPR002300 aa-tRNA-synth_Ia
IPR009008 Val/Leu/Ile-tRNA-synth_edit
IPR001412 aa-tRNA-synth_I_CS
IPR004493 Leu-tRNA-synth_Ia_arc/euk
IPR009080 tRNAsynth_Ia_anticodon-bd
IPR013155 M/V/L/I-tRNA-synth_anticd-bd
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR018114 TRYPSIN_HIS
IPR033116 TRYPSIN_SER
IPR012224 Pept_S1A_FX
IPR002300 aa-tRNA-synth_Ia
IPR009008 Val/Leu/Ile-tRNA-synth_edit
IPR001412 aa-tRNA-synth_I_CS
IPR004493 Leu-tRNA-synth_Ia_arc/euk
IPR009080 tRNAsynth_Ia_anticodon-bd
IPR013155 M/V/L/I-tRNA-synth_anticd-bd
CDD
ProteinModelPortal
H9IVN9
A0A194Q9W6
A0A1E1W615
A0A2H1WPC3
A0A2A4JX60
D2Y4U5
+ More
A0A023JNS6 B6CME7 A0A0N1IE76 I6SW41 C9W8I7 A0A2Z5U7J9 I6TRU2 A0A0L7LSM8 B6D1Q8 A0A089RSX5 I6TRT9 Q4L1L0 A0A0N1I8C7 Q9Y7A9 E9FBE3 A0A2W1BJP2 A0A2A4JTP3 A0A0L7KYH0 A0A194QET8 J7IM02 A0A194PP33 A0A0B4EQ31 B4X996 B4X9A2 B1NLD8 A0A2A4JTX8 A0A2H1WEE4 J7IN23 A0A194PVB6 A0A0N1PHJ0 A0A0M4M544 A0A0L7L163 A0A1S4H7M9 A0A2W1BE44 A0A182UPH4 Q6R560 C9W8J1 A0A2W1BTM0 A0A2W1BJK7 A0A212FL72 B4X995 O18435 A0A182LYQ9 B0ZBM9 A0A340TBF8 I7D523 J7IJ83 A0A182LPG3 A0A2H1WTV1 A0A194QDC5 A0A0B4ERM0 A0A2A4K5G7 C9W8J5 A0A0D9NL96 A0A194QJ28 C9W8I2 A0A2A4JCA8 A0A2A4K648 A0A182M5F2 A0A182KHH7 A0A2H1VGQ8 A0A395HH54 I6TMW8 A0A182MZ05 A0A182UGL3 A0A2H3FQP9 Q9NB91 Q4L1K1 Q7Q344 D3TKS0 A0A2A4JRB9 D2A2R7 A0A2H1VGP3 B6CME9 A0A182T760 Q4L1K4 C9W8F7 O18447 C9W8I6 A0A182T724 A0A194PP38 B0WBW0 J7IFG7 J3JW59 A0A2A4K636 N6TW63 U4TXR1 V9LL27 B4X9A0 A0A182YPE1 A0A2A4K6E9
A0A023JNS6 B6CME7 A0A0N1IE76 I6SW41 C9W8I7 A0A2Z5U7J9 I6TRU2 A0A0L7LSM8 B6D1Q8 A0A089RSX5 I6TRT9 Q4L1L0 A0A0N1I8C7 Q9Y7A9 E9FBE3 A0A2W1BJP2 A0A2A4JTP3 A0A0L7KYH0 A0A194QET8 J7IM02 A0A194PP33 A0A0B4EQ31 B4X996 B4X9A2 B1NLD8 A0A2A4JTX8 A0A2H1WEE4 J7IN23 A0A194PVB6 A0A0N1PHJ0 A0A0M4M544 A0A0L7L163 A0A1S4H7M9 A0A2W1BE44 A0A182UPH4 Q6R560 C9W8J1 A0A2W1BTM0 A0A2W1BJK7 A0A212FL72 B4X995 O18435 A0A182LYQ9 B0ZBM9 A0A340TBF8 I7D523 J7IJ83 A0A182LPG3 A0A2H1WTV1 A0A194QDC5 A0A0B4ERM0 A0A2A4K5G7 C9W8J5 A0A0D9NL96 A0A194QJ28 C9W8I2 A0A2A4JCA8 A0A2A4K648 A0A182M5F2 A0A182KHH7 A0A2H1VGQ8 A0A395HH54 I6TMW8 A0A182MZ05 A0A182UGL3 A0A2H3FQP9 Q9NB91 Q4L1K1 Q7Q344 D3TKS0 A0A2A4JRB9 D2A2R7 A0A2H1VGP3 B6CME9 A0A182T760 Q4L1K4 C9W8F7 O18447 C9W8I6 A0A182T724 A0A194PP38 B0WBW0 J7IFG7 J3JW59 A0A2A4K636 N6TW63 U4TXR1 V9LL27 B4X9A0 A0A182YPE1 A0A2A4K6E9
PDB
1CO7
E-value=2.28121e-19,
Score=233
Ontologies
GO
Topology
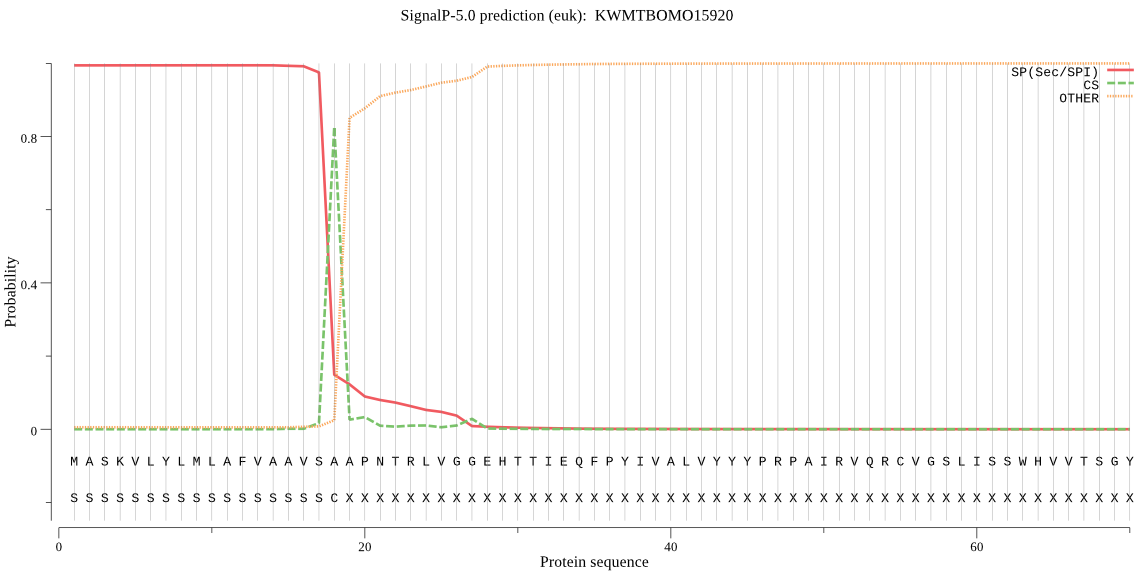
SignalP
Position: 1 - 18,
Likelihood: 0.994348
Length:
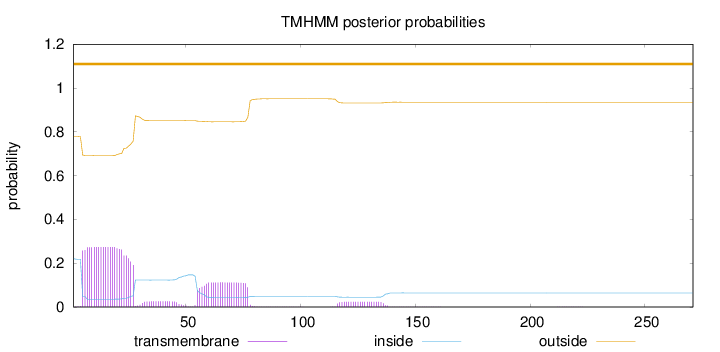
271
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.39263999999999
Exp number, first 60 AAs:
6.99452
Total prob of N-in:
0.22054
outside
1 - 271
Population Genetic Test Statistics
Pi
258.233987
Theta
204.715822
Tajima's D
-1.189387
CLR
286.978577
CSRT
0.104694765261737
Interpretation
Uncertain
