Gene
KWMTBOMO15919
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.705 Nuclear Reliability : 1.495
Sequence
CDS
ATGAACATGGACAAAACAAAAATCACGTACAATGTCCATGTTGCACCCACCATAGTTACTATTGGGAGCTCTACTCTTGAAGTTGTTGACGAATATATATACCTAGGACAGGCAGTCCGATTATGTAGGTCCAACTTTGAGAGAGAGGTCAGTAGACGGATCCAACTCGGCTGGGCAGCGTTCGGTAAACTGCACAACGTCTTCTCGTCCAAAATACCTCAGTGCCTTAAGACAAAGGCGTACAACCGATGTGTGTTACCAGTGATGACTTACGGCTCGGAGACGTGGTGCTTCACTAGGGATCTTTTTAGAAGGCTCAAAGTCGCGCAGCGAGCTATGGAGAGGGCTATGCTCGGCATTTCTCTACGTGATCGAATTCGAAATGAGGAGATCCGTAGAAGAACGAAGGTTGCTGACATAGCCCGTATAATTAGTGAACTGAAGTGGCAATGGGCCGGCCACATAGCACGAAGAGAGGACGGTCAATGGGGCAGAAAGATCCTGGAGTGGCGACCACGTACTGGCAAGCGCAGTGTGGGACGGCCACCTACTAGATGGACCGACGACCTAGTAAAAAGCGCTGGGTCTCGTTGGATGCAGGTGGCAATGGACCGGTCACACTGGAAATCTATTGGAAAGGCCTATGTTCAGCAGTGGACGGCGATAGGCTGA
Protein
MNMDKTKITYNVHVAPTIVTIGSSTLEVVDEYIYLGQAVRLCRSNFEREVSRRIQLGWAAFGKLHNVFSSKIPQCLKTKAYNRCVLPVMTYGSETWCFTRDLFRRLKVAQRAMERAMLGISLRDRIRNEEIRRRTKVADIARIISELKWQWAGHIARREDGQWGRKILEWRPRTGKRSVGRPPTRWTDDLVKSAGSRWMQVAMDRSHWKSIGKAYVQQWTAIG
Summary
Uniprot
D7F158
D7F159
A4KWF4
A0A3S2TAR4
A4KWF2
A4KWG0
+ More
D7F164 A0A2W1BMB6 D7F157 D7F166 D7F165 D7F160 A0A3S2N4D5 A0A2H1WG43 A0A3S2KX72 A0A2W1BLC9 A0A3S2TRE0 A0A2G8JIM1 W4YXJ1 A0A1E1X2U1 A0A2G8JFM9 A0A131YK20 A0A023EXV2 A0A2G8KFR2 A0A2W1BCS2 E2AL50 A0A2G8K4F2 D7F178 A0A0P4VWD0 D7F176 A0A069DX96 D7F177 A0A023EWU8 A0A0M3HSL8 A0A183UMK8 A0A2G8KKE1 A0A2A4J310 A0A2A4J025 D7F179 A0A016W5P9 A0A016VZE9 A0A016VZ67 A0A016SG46 A0A2G8KAA6 A0A016TK11 A0A016W4R1 A0A069DQ34 A0A016V419 A0A016S6G1 A0A2W1B5F8 A0A016VHV5 A0A016UHF7 A0A2G8KES6 A0A016U3R5 A0A016W591 A0A016TIA8 E3MII9 A0A016WEQ1 E3NMS2 A0A016TNT0 A0A2W1BSQ8 A0A016S5B2 A0A016WA79 A0A2H1VY30 E3NG80 A0A016S795 A0A2W1BDX2 A0A224XSN7 F1LDF6 A0A016TPB0 E3NQY5 A0A016VVN3 E3NNK9 A0A016T7J7 A0A2H2JCJ4 E3NCZ1 A0A2G8L8R5 E3NSE7 A0A2H1V663 E3NBP3 A0A016U784 A0A016S403 E3MW49 E3MYM4 E3N7N4 A0A2W1BZV1 A0A016TS31
D7F164 A0A2W1BMB6 D7F157 D7F166 D7F165 D7F160 A0A3S2N4D5 A0A2H1WG43 A0A3S2KX72 A0A2W1BLC9 A0A3S2TRE0 A0A2G8JIM1 W4YXJ1 A0A1E1X2U1 A0A2G8JFM9 A0A131YK20 A0A023EXV2 A0A2G8KFR2 A0A2W1BCS2 E2AL50 A0A2G8K4F2 D7F178 A0A0P4VWD0 D7F176 A0A069DX96 D7F177 A0A023EWU8 A0A0M3HSL8 A0A183UMK8 A0A2G8KKE1 A0A2A4J310 A0A2A4J025 D7F179 A0A016W5P9 A0A016VZE9 A0A016VZ67 A0A016SG46 A0A2G8KAA6 A0A016TK11 A0A016W4R1 A0A069DQ34 A0A016V419 A0A016S6G1 A0A2W1B5F8 A0A016VHV5 A0A016UHF7 A0A2G8KES6 A0A016U3R5 A0A016W591 A0A016TIA8 E3MII9 A0A016WEQ1 E3NMS2 A0A016TNT0 A0A2W1BSQ8 A0A016S5B2 A0A016WA79 A0A2H1VY30 E3NG80 A0A016S795 A0A2W1BDX2 A0A224XSN7 F1LDF6 A0A016TPB0 E3NQY5 A0A016VVN3 E3NNK9 A0A016T7J7 A0A2H2JCJ4 E3NCZ1 A0A2G8L8R5 E3NSE7 A0A2H1V663 E3NBP3 A0A016U784 A0A016S403 E3MW49 E3MYM4 E3N7N4 A0A2W1BZV1 A0A016TS31
Pubmed
EMBL
FJ265543
ADI61811.1
FJ265544
ADI61812.1
EF113399
EF113400
+ More
ABO45233.1 RSAL01002362 RVE40503.1 EF113398 EF113401 ABO45231.1 EF113402 ABO45239.1 FJ265549 ADI61817.1 KZ150072 PZC74016.1 FJ265542 ADI61810.1 FJ265551 ADI61819.1 FJ265550 ADI61818.1 FJ265545 ADI61813.1 RSAL01000839 RVE41117.1 ODYU01008431 SOQ52031.1 RSAL01002130 RVE40575.1 KZ149985 PZC75698.1 RSAL01000016 RVE53061.1 MRZV01001865 PIK35593.1 AAGJ04144474 GFAC01005640 JAT93548.1 MRZV01002136 PIK34564.1 GEDV01010296 JAP78261.1 GBBI01004876 JAC13836.1 MRZV01000618 PIK46838.1 KZ150485 PZC70710.1 GL440455 EFN65840.1 MRZV01000899 PIK42833.1 FJ265563 ADI61831.1 GDKW01001596 JAI54999.1 FJ265561 ADI61829.1 GBGD01000453 JAC88436.1 FJ265562 ADI61830.1 GBBI01004877 JAC13835.1 UYWY01020264 VDM41049.1 MRZV01000520 PIK48481.1 NWSH01003335 PCG66527.1 NWSH01004509 PCG65028.1 FJ265564 ADI61832.1 JARK01000732 EYC35149.1 JARK01001338 EYC32701.1 EYC32700.1 JARK01001565 EYB89683.1 MRZV01000743 PIK44934.1 JARK01001676 JARK01001446 JARK01001433 EYB83199.1 EYC01126.1 EYC02928.1 JARK01001337 EYC34625.1 GBGD01002934 JAC85955.1 JARK01001354 EYC21986.1 JARK01001626 EYB85834.1 KZ150317 PZC71439.1 JARK01001346 EYC26368.1 JARK01001376 JARK01001370 JARK01001342 EYC14570.1 EYC16380.1 EYC29217.1 MRZV01000640 PIK46507.1 JARK01001396 EYC09581.1 JARK01000807 EYC34984.1 JARK01001434 EYC02684.1 DS268448 EFP02560.1 JARK01000347 EYC38051.1 DS269097 EFP08676.1 JARK01001424 EYC04322.1 KZ149912 PZC78089.1 EYB85833.1 JARK01000460 EYC36749.1 ODYU01004982 SOQ45402.1 DS268650 EFO96957.1 JARK01001621 EYB86109.1 KZ150108 PZC73382.1 GFTR01005437 JAW10989.1 JI178870 ADY48160.1 EYC04323.1 DS269613 EFO86468.1 JARK01001340 EYC30828.1 DS269225 EFP10829.1 JARK01001736 JARK01001462 JARK01001379 EYB80775.1 EYB98943.1 EYC13646.1 EYC33347.1 DS268603 EFO93378.1 MRZV01000169 PIK56634.1 DS269943 EFO90109.1 ODYU01000693 SOQ35892.1 DS268587 EFO92044.1 JARK01001389 EYC11015.1 JARK01001639 EYB85201.1 DS268485 EFP10420.1 DS268497 EFP12149.1 DS268549 EFO89066.1 KZ149907 PZC78256.1 JARK01001415 EYC05859.1
ABO45233.1 RSAL01002362 RVE40503.1 EF113398 EF113401 ABO45231.1 EF113402 ABO45239.1 FJ265549 ADI61817.1 KZ150072 PZC74016.1 FJ265542 ADI61810.1 FJ265551 ADI61819.1 FJ265550 ADI61818.1 FJ265545 ADI61813.1 RSAL01000839 RVE41117.1 ODYU01008431 SOQ52031.1 RSAL01002130 RVE40575.1 KZ149985 PZC75698.1 RSAL01000016 RVE53061.1 MRZV01001865 PIK35593.1 AAGJ04144474 GFAC01005640 JAT93548.1 MRZV01002136 PIK34564.1 GEDV01010296 JAP78261.1 GBBI01004876 JAC13836.1 MRZV01000618 PIK46838.1 KZ150485 PZC70710.1 GL440455 EFN65840.1 MRZV01000899 PIK42833.1 FJ265563 ADI61831.1 GDKW01001596 JAI54999.1 FJ265561 ADI61829.1 GBGD01000453 JAC88436.1 FJ265562 ADI61830.1 GBBI01004877 JAC13835.1 UYWY01020264 VDM41049.1 MRZV01000520 PIK48481.1 NWSH01003335 PCG66527.1 NWSH01004509 PCG65028.1 FJ265564 ADI61832.1 JARK01000732 EYC35149.1 JARK01001338 EYC32701.1 EYC32700.1 JARK01001565 EYB89683.1 MRZV01000743 PIK44934.1 JARK01001676 JARK01001446 JARK01001433 EYB83199.1 EYC01126.1 EYC02928.1 JARK01001337 EYC34625.1 GBGD01002934 JAC85955.1 JARK01001354 EYC21986.1 JARK01001626 EYB85834.1 KZ150317 PZC71439.1 JARK01001346 EYC26368.1 JARK01001376 JARK01001370 JARK01001342 EYC14570.1 EYC16380.1 EYC29217.1 MRZV01000640 PIK46507.1 JARK01001396 EYC09581.1 JARK01000807 EYC34984.1 JARK01001434 EYC02684.1 DS268448 EFP02560.1 JARK01000347 EYC38051.1 DS269097 EFP08676.1 JARK01001424 EYC04322.1 KZ149912 PZC78089.1 EYB85833.1 JARK01000460 EYC36749.1 ODYU01004982 SOQ45402.1 DS268650 EFO96957.1 JARK01001621 EYB86109.1 KZ150108 PZC73382.1 GFTR01005437 JAW10989.1 JI178870 ADY48160.1 EYC04323.1 DS269613 EFO86468.1 JARK01001340 EYC30828.1 DS269225 EFP10829.1 JARK01001736 JARK01001462 JARK01001379 EYB80775.1 EYB98943.1 EYC13646.1 EYC33347.1 DS268603 EFO93378.1 MRZV01000169 PIK56634.1 DS269943 EFO90109.1 ODYU01000693 SOQ35892.1 DS268587 EFO92044.1 JARK01001389 EYC11015.1 JARK01001639 EYB85201.1 DS268485 EFP10420.1 DS268497 EFP12149.1 DS268549 EFO89066.1 KZ149907 PZC78256.1 JARK01001415 EYC05859.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
D7F158
D7F159
A4KWF4
A0A3S2TAR4
A4KWF2
A4KWG0
+ More
D7F164 A0A2W1BMB6 D7F157 D7F166 D7F165 D7F160 A0A3S2N4D5 A0A2H1WG43 A0A3S2KX72 A0A2W1BLC9 A0A3S2TRE0 A0A2G8JIM1 W4YXJ1 A0A1E1X2U1 A0A2G8JFM9 A0A131YK20 A0A023EXV2 A0A2G8KFR2 A0A2W1BCS2 E2AL50 A0A2G8K4F2 D7F178 A0A0P4VWD0 D7F176 A0A069DX96 D7F177 A0A023EWU8 A0A0M3HSL8 A0A183UMK8 A0A2G8KKE1 A0A2A4J310 A0A2A4J025 D7F179 A0A016W5P9 A0A016VZE9 A0A016VZ67 A0A016SG46 A0A2G8KAA6 A0A016TK11 A0A016W4R1 A0A069DQ34 A0A016V419 A0A016S6G1 A0A2W1B5F8 A0A016VHV5 A0A016UHF7 A0A2G8KES6 A0A016U3R5 A0A016W591 A0A016TIA8 E3MII9 A0A016WEQ1 E3NMS2 A0A016TNT0 A0A2W1BSQ8 A0A016S5B2 A0A016WA79 A0A2H1VY30 E3NG80 A0A016S795 A0A2W1BDX2 A0A224XSN7 F1LDF6 A0A016TPB0 E3NQY5 A0A016VVN3 E3NNK9 A0A016T7J7 A0A2H2JCJ4 E3NCZ1 A0A2G8L8R5 E3NSE7 A0A2H1V663 E3NBP3 A0A016U784 A0A016S403 E3MW49 E3MYM4 E3N7N4 A0A2W1BZV1 A0A016TS31
D7F164 A0A2W1BMB6 D7F157 D7F166 D7F165 D7F160 A0A3S2N4D5 A0A2H1WG43 A0A3S2KX72 A0A2W1BLC9 A0A3S2TRE0 A0A2G8JIM1 W4YXJ1 A0A1E1X2U1 A0A2G8JFM9 A0A131YK20 A0A023EXV2 A0A2G8KFR2 A0A2W1BCS2 E2AL50 A0A2G8K4F2 D7F178 A0A0P4VWD0 D7F176 A0A069DX96 D7F177 A0A023EWU8 A0A0M3HSL8 A0A183UMK8 A0A2G8KKE1 A0A2A4J310 A0A2A4J025 D7F179 A0A016W5P9 A0A016VZE9 A0A016VZ67 A0A016SG46 A0A2G8KAA6 A0A016TK11 A0A016W4R1 A0A069DQ34 A0A016V419 A0A016S6G1 A0A2W1B5F8 A0A016VHV5 A0A016UHF7 A0A2G8KES6 A0A016U3R5 A0A016W591 A0A016TIA8 E3MII9 A0A016WEQ1 E3NMS2 A0A016TNT0 A0A2W1BSQ8 A0A016S5B2 A0A016WA79 A0A2H1VY30 E3NG80 A0A016S795 A0A2W1BDX2 A0A224XSN7 F1LDF6 A0A016TPB0 E3NQY5 A0A016VVN3 E3NNK9 A0A016T7J7 A0A2H2JCJ4 E3NCZ1 A0A2G8L8R5 E3NSE7 A0A2H1V663 E3NBP3 A0A016U784 A0A016S403 E3MW49 E3MYM4 E3N7N4 A0A2W1BZV1 A0A016TS31
Ontologies
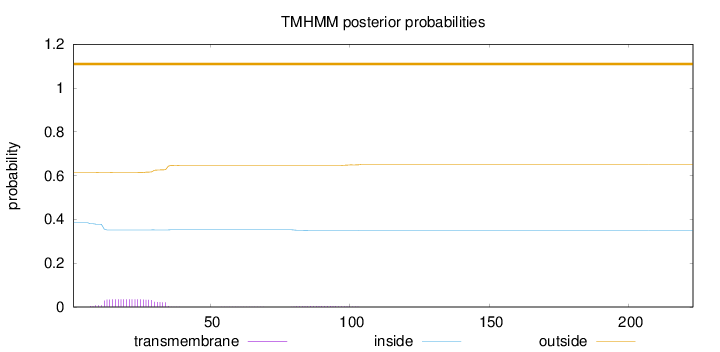
Topology
Length:
223
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.82757
Exp number, first 60 AAs:
0.75605
Total prob of N-in:
0.38513
outside
1 - 223
Population Genetic Test Statistics
Pi
144.147606
Theta
108.195247
Tajima's D
0.782587
CLR
0
CSRT
0.60121993900305
Interpretation
Uncertain
