Pre Gene Modal
BGIBMGA013415
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC101738244_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.897
Sequence
CDS
ATGGAAAACATGTCTTCGAATGTGTCTCTACGGAATAAACAGAGGAAAGATAATCTTATCACAAGGAAACAGGATAAGTCGCCGGAAAAGACCATCAGAGGAACTAGGGGAGCCTACAGGGCGGTCCCTTCCACTTCGAAGAGGAACGATGAGGAGGAAACGGGCTCCACGGAAAGACTGGACAGACCAAAGAAATTCATATCTCAGAATAAAACCAACTACGGTCTGGGGAAGAGGCCAGACTTCTTGCTTAAGAACAGGAATGTGCATTTAGCGTCCTCTCGCTATGGACAGATGACCTCTGGGCTTCACGGGCACGGCCACCAGATCCCGCAGCCTCCCGCGCACCATCTAGACCCCGCCGCTGTGATCCCCACCAAGGAGAAAACAATACACGGGAAGGAAAAGACGTTTAAAGTCGTGGTGGGCGGAGGGGAGGTCGGCGTGATGAAGGCTCCGGTCCTGGGGCCGAGGCCGTACAATGCGCTCGCCATCATCCATACTCAGCAGAGCAACACCTTTGATATGCTCAACTCGATGACCACAAGCGTAGCGAGTCAGCCCGTGGTAGGCGGGTTCCGCCGGCGTAGCTCGGGTCAGGGCGCTCTCTGCGACCCTGAGCTGGACGACAGCGGGAACAAGCTGGTCGCCAGGCTGGAGGACGACGCCTTCATATCCGAACAGGATCTGGAAGAGAACATCCAGATAGCGACCGGCACGCAGGAGGGTCTCCCCCGCCACGGCGCCGCCGCTGATGAGGGGCAGCCCCAAGAGCCTCAGAACGGTCTAGAATACGGTCCTATTTACGAGCTGGAGAAGCTCGAAGAGAAAGACAAACAGAAGCAAGACAAGGAAGATGAGGAGGAGCCGATCGGAGTATCACCGTGCGGCAGGTTTTTCAAATACGACAAGGAAGTGGGTCGCGGTTCGTTCAAGACCGTCTACCACGGACTCGACACCCAGACCGGCGTCGCCGTTGCCTGGTGCGAGTTACTCGAGAAAAAATTGAACAAGACGGAGCGTCTGCGTTTCCGCGAGGAGGCCGACATGCTGAAGAAGCTGCAGCACCCGAACATCGTCCGCTTCTACAACTACTGGGAGGGGACCGTCGCCAAGAAGAAGAACATCGTGCTCATCACCGAGCTCATGCTCTCCGGGACGCTCAAGGCCTACCTCAGGAGATTCAAGCGAATCAACCCGAAGGTGTTGAAGTCGTGGTGTCGCCAAATACTCAAGGGTCTGAATTTTTTACATTCACGAACTCCGCCTATAATACACAGAGATCTGAAGTGCGACAACATCTTCATCACCGGCACGACGGGCAGCGTCAAGATCGGGGACCTCGGCCTCGCCACGCTCAAGAATAGGAGCTTCGCCAAGTCTGTCATCGGCACACCCGAGTTCATGGCGCCGGAGATGTACGAGGAGCACTACGACGAGTCCGTGGACGTGTACGCGTTCGGCATGTGCATGCTCGAGATGGCCACCGGCGAGTACCCCTACAGCGAGTGCAACAGCCCCGCGCAGATCTACAAGAAGGTCGTCTCGGGCGTGAAGCCACAGAGCTTGGAGAAGGTCACCATCCCCGAGGTGAGGGAGATCATCGAGTCGTGCATAAAGCCGAATAAAGTTGATCGGCCAAAAGTGAAAGATCTGCTAAACCACGAGTTCTTCGGCGAAGACATTGGACTTAGATTGGAGATAGTCGACCGAGACCTGGTCACGTCCTCGGACGTAACCAAAATACAGTTCCGTCTCAAGATCATCGACCCCAAAAAACGGTCGTACACGCACAAAGAGAACGAAGCGATACAGTTTGAGTTCGACGTGGAGCACGACGACTGCGAGGAGGTGGCCAACGAGATGGCCAAGACCGGCCTCATCATGGAGGAGGACACCAGAATCGTCTCCAAGCTGCTGAAATCGCAACTCATTAGCTTAAATCGTGAACGGACCGAGAAGAAAGCTCAACTTTTGTTCAACCAAGAAGTCATGATCGAACAGAATCGACAACAACAGATGTTTGAGCAACAACTTCGACAGGTGAGTTGA
Protein
MENMSSNVSLRNKQRKDNLITRKQDKSPEKTIRGTRGAYRAVPSTSKRNDEEETGSTERLDRPKKFISQNKTNYGLGKRPDFLLKNRNVHLASSRYGQMTSGLHGHGHQIPQPPAHHLDPAAVIPTKEKTIHGKEKTFKVVVGGGEVGVMKAPVLGPRPYNALAIIHTQQSNTFDMLNSMTTSVASQPVVGGFRRRSSGQGALCDPELDDSGNKLVARLEDDAFISEQDLEENIQIATGTQEGLPRHGAAADEGQPQEPQNGLEYGPIYELEKLEEKDKQKQDKEDEEEPIGVSPCGRFFKYDKEVGRGSFKTVYHGLDTQTGVAVAWCELLEKKLNKTERLRFREEADMLKKLQHPNIVRFYNYWEGTVAKKKNIVLITELMLSGTLKAYLRRFKRINPKVLKSWCRQILKGLNFLHSRTPPIIHRDLKCDNIFITGTTGSVKIGDLGLATLKNRSFAKSVIGTPEFMAPEMYEEHYDESVDVYAFGMCMLEMATGEYPYSECNSPAQIYKKVVSGVKPQSLEKVTIPEVREIIESCIKPNKVDRPKVKDLLNHEFFGEDIGLRLEIVDRDLVTSSDVTKIQFRLKIIDPKKRSYTHKENEAIQFEFDVEHDDCEEVANEMAKTGLIMEEDTRIVSKLLKSQLISLNRERTEKKAQLLFNQEVMIEQNRQQQMFEQQLRQVS
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
PDB
5O2C
E-value=6.09782e-140,
Score=1277
Ontologies
GO
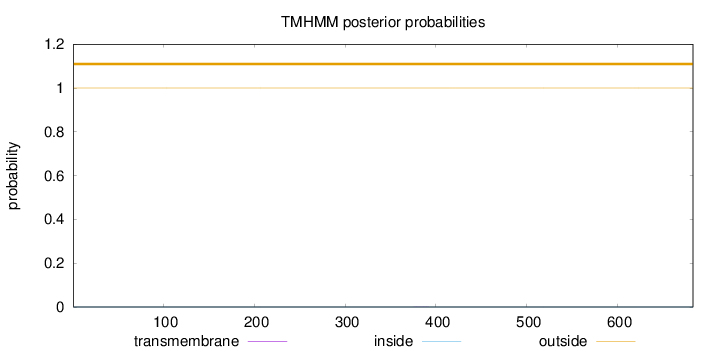
Topology
Length:
683
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00536
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00031
outside
1 - 683
Population Genetic Test Statistics
Pi
22.136533
Theta
16.06658
Tajima's D
0.465055
CLR
1364.813554
CSRT
0.515374231288436
Interpretation
Uncertain
