Pre Gene Modal
BGIBMGA013394
Annotation
PREDICTED:_protein_MON2_homolog_[Amyelois_transitella]
Full name
Protein MON2 homolog
Alternative Name
Protein SF21
Location in the cell
Cytoplasmic Reliability : 1.107 PlasmaMembrane Reliability : 1.489
Sequence
CDS
ATGGCATTCGTGAGTGCCGTAACCGGAGATGATTCAACAAAGAAGTTCATGGAAGTCCTTCAGAACGACTTTAAAAATTTAAGTTTGGAGACCAAGAAGAAATATCCTCAGATAAGAGAGGCTTGCGATGAAGCTATAGAAAAACTGTCTTTAGCCTCTAATAATCCACAAGCTTCACTGTATGGTGTTGTTAATCAAATATTGTATCCACTGGTGCAAGGTTGTGAATCAAAAGATTTGAAAATTATTAAGTTCTGCCTAGGGACTATCCAGCGGTTAATAGCTCAGCAAGGGATAGACGCGAAAGGCGCAAGGCATGTGGTGGACTGCTTGTACAATCTAGGCCAGGCCGGGGTCTTAGAGCTAAAATTGCTTCAAACTGCTGCTCTGTTGATGACTACCTCTGATTTGGTACATGGAGACACGTTGGCACGGACAATGGTGATGTGTATGAAGATGGTATCCACAACGGAGACAAGGGATATTAGCACGAGTCACGCCGCAGCCGCCACTGTGAGGCAACTGGTCGCCTTAGTTTTTGAGAGGGCACTTGCTGAGGCAGATGGTAATCTAAAAGTGAATCCGGCCGATGTTAGATTGCAGAGTAATAACAAAGCGCCAAAGGACCTCAAACCGTGTGCCGTTGATGCCTATCTTATTTTGCAAGATATAATTCAGTTAATAAACGGTGACGCCGCCCACTGGCTCGTCGGCATATCGGATGTGCCCAAGACATTCGGTCTGGAGTTACTGGACACAGTACTTACTGACTTCTCACCGGTTTTCTTCAAGATATCCGAGTTCAGGTTCCTGTTGAAGGAGCACGTTTGCGCGCTGATAATCAAACTGTTCTCACCGAACGTCAAGTACAGGGCTGCGTTGACGGCATTGCACATCCCGGGGGCTGCGGGCGCGAACGGGGGCGGAACACCCGAGCGCCCGCACTTCCCCGTCACGATGAGGCTACTCAGACTCGTCTCAGTAATTCTACACAAATACCACTCTTTACTTGTGACGGAGTGCGAAATATTTCTGTCGCTGACGATAAAATTTCTAGATCCCGACAAGCCGTTGTGGCAGCGAGCTTTAGCTCTAGAAGTTTTACACAAGATGACAATACAGCCAAAGCTGCTGAAGTCGTTCTGCGACTGCTACGACATGAAGCCGCACGCCACCAACATCTTCCAGGACATCGTGAACGCGCTGGGCGCCTACGTGCAGAGCCTGTTCGCGTCACCAGGAGCGGCCGCGCCCCCCGGTACATCGGCGATACCTCAGCAATCGGGTTTCTACTGCAAAGGAGTTTGGCTGCCACTCTGCGTCACATTCGAGCGTGGTACAGCCAAGGCTGTCTACATAGAGATGCTGGATCGAACCGAGGCCCCCACGATCCAAGACGGATACGGCATCTCCGTCGCCTACGCATGCTTGATGGAAATAATACGGTCCATAGCCATAACCGTGGAGGGGTCCGAGTACTTCAGGTTACAAGAAATGTATGAAGATCTTCATGACGGCGATGAAAAAGAACTCAATTCAAATAAAACTAACACAGACACGGCCACGACAGAGCAGAGCTTAACCAGTAACGGGCACAGCGTCGAAGCAGATCAGAATTCGAACGTAGTTCAAGCGGCCTCTGACGATAGCGACAAAGAAACGGAGCTGAGATTACAGTTGTTAAAGTCTTCGTGGTGCGGCCTGGTGTGGGGCTTGTCAGTGCTGGCTGACGCGAGCATCGGCGAGCTGGAGAACATACTGCGCGCCATACAGACGCTGGCCAGGGTCAGCGGCAAGGTGGGGGTGTCTGCGGGGCGGGACGCGTGCGTGGGCGCGTTGTGCCGCAGCGCGCTGCCCGCGCAGTACGCCGCCGGAGCCCTGGGCGCGTGCCCGTGGAGACGCGAGCCCGCCGCGCCCGAGCCCGACTGCCGGCACCACGTCGTGTGGGTCGGGACGCCGCTGCCCTCGGCCGCGCCGGCCTCTGGACAGCAGCAGTCTTTCGTGATGGTGACGTCAAGACACGTGACCGCCATGAAGGCGCTACTGTGCGCGGCGCAGCGCGACGGTGACGTCATACAGCAGGCCTGGTTCCCGGTTCTAACCACGCTGCAGCATTTGGTCTGGATCTTGGGCCTGAAGCCGTCGACTGGCGGCAGTATGAAGGCCAGTCGCGCCAGTGCAGACGCCAACGCGGTCATGAGCACGTCCGCGGTAATGGCAGACTTGCCCGCTCTGTCAGCAATGCTCTCCCGGGTGTTCGAGTCTTCCAAGGACTTGGACGACGTGGCTCTGCACCACCTAATCGACGCGCTGTGCAAGCTCTCCAACGAGGCCATGGAACTGGCTTATTCAAATCGGGAGCCATCGCTGTTCGCCGTGGCAAAACTTCTAGAAACAGGTCTGGCGAACATGCACCGGATAGAGGTGATGTGGAGGCCCATCACCAACCACTTGTTGGAGGTGTGTCAGCACCCGCACATCAGGATGCGGGAATGGGGAGTGGAGGCCATCACGTATCTAGTTCAAGCAGCATTCCAGTACCACCATAATAATCCCGTTCATGTAACTGATGCCCGGTCCCGGCTGCTGCTGGAGCCGCTGGCGGAGCTGTGCTCGGTGCGGCACGCGGACGTGCGCGCCCGCCAGCTGGAGTGCGCGGCGCGGCTGCTGCACTCGCGCGGGGACCAGCTGGGCGCCGCCTGGCCGCTCATGATGCAAGTCATCTCCGCCATCTCCGAGCACCACAGCGAGCAGCTAATCAGGTCGGCGTTCCAGTGCGCGCAGGTGGTGGCCGGCGACCTGCTGGGCTGCGCCGGCCCGCGCTGCCTGCGCCGCGTGCTCAACACGGCCGCCGCCTTCGCGCTGCAGACCAAGGAGCTCAACATCAGCCTCACCGCCGTGGGACTCATGTGGAACATATCGGACTACCTGTACCACAACCGCGACAAGCTGGTGGCGGCGCTGGCGGGCGACGCGGCCGCGTGTCCCGAGGCGCCGCAGCACGACGAGCTGCCGCCGCTCGACCGCCTCTGGATGTGCCTCTACATCAGACTTAGCGAGCTGTGCACGGAGCCCCGCGCCCCGGTCCGGCGCGCCGCCAGCCAGACGCTGTTCAGCTGCATCGGCGCGCACGGGGCGCTGCTCGGCAAGCCCGCCTGGCGCGCCCTGCTCGCCGTGCTCTTCCCCATGCTCGAGCAGGTCCAGAAACAATCAAACATTGCGAGCTGCGAGAAGGTGGACACCGGGGAGCACATCCTCATACATCACACGAGAAACACGGCGCAGAAACAGTGGGCGGAGACCCAGGTGCTCACGTTATCGGGGGTGTCGAGAGTCTTCCACTCCCGATTCCAGCTCCTGACGACCGTGGGAGACTTCAACAGATCCTGGGCGATCCTGCTGGGCTACATAACCGACTTCGCCTTACAGAAGAGTCACGAGGTGTCGGTGGCGGCGCTGAAGTCGTTCCAGGAGGTGGTGTCGGCGGCGGGGCGCGCGGGGGGGCTGCCGCGCCGCGTGTGGGACGCCGCGTGGGCCGCCTGGACCGACATCGCCGCGGCGCTGCCGGACCGACACCGCGAGCCCCCGGTGACGGAGGACGGCAAGCCAGGCGAGGAGTACGCGCCCTCCCAGAGCTTCCTCACGACCCTCGTGCAAATATTCCCGCTTATATTCCAGCACATACGGCCCACGTTCACGGCGGCGGACGTGGAGCGGCTGGGCGCGTGCCTGGCGCACGTGTGCCGCATCGAGCCCTCCGCCTCGGCCTCCGCCCTGGACGGGTCCGCCGGGCTCGCCGTCAGCCTGCACGCCCTGCACTGCCTCGACACCGTGCACAAGGAGGCGCTGGTCCGGCACGAGCTGCTGGGCGCGATGTTCGTGGCGCTGACGTCACTGGCGGGCGCGGGCGCGGGTGCGGGCGCGCGCTGCCTGTCCGCCGCCGTGGCGCTGTACCGGGCCGCGCCGCAGCCCAGCGCCCCCGTGCTGCCGCATCTCTTGCAGGCTCTGCACTCGGCGGTCAAGAAGTGCGGGCAGGAGAAACGTCGCAGACCCTCGGAACAAGTGGACGAGGCGTCGCAAATAGCGGCCCTATTACTACAGGTATTGGAAACCGGTCTACCACTAGCGAGAGAAAAGCCAGACGACTACAAAGAGTTCTGGGAGACGCTTCCCGGAGTGCTCGAGACGTTCCTGTTTGAGCCACCTACGGGGATGGGCGCGGGTGCGGGCGCGCTCGTGGCGTGCGTGCGCGGCGCGGCGCTGGCGGGGCGCGTGCCGGGCGCGGCGCTGGGCCGCCTGCTGGCGCTGGTGCGGGCCGCCGCGCTGCACCACGCGCCGCCGGACCGCACGCAGACCGACCAAGAACTGAAAGAGAGGGAAGAATTCGCCCGGACGTGCTTCGAGACGCTACTGCAGTTCTCTATGGTGGAGAACGTCGACTCCATTTCCGGGGATGACAATGATTCTGACCCGCTAGCCATAATGACGTTGCTGGACAGGTTCCAAGAGATCATAATGAAGTATGCAGAGGATGAGCACGACACAGAGCCTCTGCCAAGACATCAACTATCGGAGATATCGTTTGTACTGAAAGCCATAGCGACGCTGGCGGAGTCCATGAAGAAAGCGCCCCCCGGGAAAGTGGACGCGGCGGCCTGGCGGAAGTTAATAGAATAG
Protein
MAFVSAVTGDDSTKKFMEVLQNDFKNLSLETKKKYPQIREACDEAIEKLSLASNNPQASLYGVVNQILYPLVQGCESKDLKIIKFCLGTIQRLIAQQGIDAKGARHVVDCLYNLGQAGVLELKLLQTAALLMTTSDLVHGDTLARTMVMCMKMVSTTETRDISTSHAAAATVRQLVALVFERALAEADGNLKVNPADVRLQSNNKAPKDLKPCAVDAYLILQDIIQLINGDAAHWLVGISDVPKTFGLELLDTVLTDFSPVFFKISEFRFLLKEHVCALIIKLFSPNVKYRAALTALHIPGAAGANGGGTPERPHFPVTMRLLRLVSVILHKYHSLLVTECEIFLSLTIKFLDPDKPLWQRALALEVLHKMTIQPKLLKSFCDCYDMKPHATNIFQDIVNALGAYVQSLFASPGAAAPPGTSAIPQQSGFYCKGVWLPLCVTFERGTAKAVYIEMLDRTEAPTIQDGYGISVAYACLMEIIRSIAITVEGSEYFRLQEMYEDLHDGDEKELNSNKTNTDTATTEQSLTSNGHSVEADQNSNVVQAASDDSDKETELRLQLLKSSWCGLVWGLSVLADASIGELENILRAIQTLARVSGKVGVSAGRDACVGALCRSALPAQYAAGALGACPWRREPAAPEPDCRHHVVWVGTPLPSAAPASGQQQSFVMVTSRHVTAMKALLCAAQRDGDVIQQAWFPVLTTLQHLVWILGLKPSTGGSMKASRASADANAVMSTSAVMADLPALSAMLSRVFESSKDLDDVALHHLIDALCKLSNEAMELAYSNREPSLFAVAKLLETGLANMHRIEVMWRPITNHLLEVCQHPHIRMREWGVEAITYLVQAAFQYHHNNPVHVTDARSRLLLEPLAELCSVRHADVRARQLECAARLLHSRGDQLGAAWPLMMQVISAISEHHSEQLIRSAFQCAQVVAGDLLGCAGPRCLRRVLNTAAAFALQTKELNISLTAVGLMWNISDYLYHNRDKLVAALAGDAAACPEAPQHDELPPLDRLWMCLYIRLSELCTEPRAPVRRAASQTLFSCIGAHGALLGKPAWRALLAVLFPMLEQVQKQSNIASCEKVDTGEHILIHHTRNTAQKQWAETQVLTLSGVSRVFHSRFQLLTTVGDFNRSWAILLGYITDFALQKSHEVSVAALKSFQEVVSAAGRAGGLPRRVWDAAWAAWTDIAAALPDRHREPPVTEDGKPGEEYAPSQSFLTTLVQIFPLIFQHIRPTFTAADVERLGACLAHVCRIEPSASASALDGSAGLAVSLHALHCLDTVHKEALVRHELLGAMFVALTSLAGAGAGAGARCLSAAVALYRAAPQPSAPVLPHLLQALHSAVKKCGQEKRRRPSEQVDEASQIAALLLQVLETGLPLAREKPDDYKEFWETLPGVLETFLFEPPTGMGAGAGALVACVRGAALAGRVPGAALGRLLALVRAAALHHAPPDRTQTDQELKEREEFARTCFETLLQFSMVENVDSISGDDNDSDPLAIMTLLDRFQEIIMKYAEDEHDTEPLPRHQLSEISFVLKAIATLAESMKKAPPGKVDAAAWRKLIE
Summary
Description
May be required for traffic between late Golgi and early endosomes.
Similarity
Belongs to the MON2 family.
Keywords
Acetylation
Alternative splicing
Complete proteome
Phosphoprotein
Polymorphism
Protein transport
Reference proteome
Transport
Feature
chain Protein MON2 homolog
splice variant In isoform 3.
sequence variant In dbSNP:rs10219555.
splice variant In isoform 3.
sequence variant In dbSNP:rs10219555.
Uniprot
A0A3S2NZ94
A0A212EJV0
A0A2J7RNE8
A0A026WRE2
A0A3L8DX75
A0A151X6D3
+ More
A0A151IH52 F4WH85 A0A195F3L0 A0A195B9D3 E2BVY3 A0A195E5V5 A0A158NPD6 A0A088AHQ3 A0A2A3E4W7 A0A154P146 K7J2C6 A0A0J7L4K9 A0A232FB04 A0A1B6LWA5 A0A1B6KIN6 A0A1Y1M6D5 A0A1W4X2P8 A0A0C9RWJ8 D6W829 A0A310SFV2 A0A0L7RES7 E2AXI0 A0A1L8DV77 E0VBS7 U4TUS9 N6TSW3 A0A0M9ABM2 A0A2M4B960 A0A2M4B8J4 A0A1B6CWH9 A0A0K8UL09 W8C084 A0A034V2P6 A0A1A9ZEU2 A0A0T6AXK1 A0A1A9XUY6 A0A1B0ASB9 A0A0N7ZAU2 A0A1B0FP44 A0A1A9UW08 A0A3R7NKU3 A0A0M4EPF9 M3YZ50 W5P2N3 A0A2Y9KYG8 U3DXG9 A0A2Y9L534 U6DIP7 A0A1S2ZBS3 A0A1S2ZBQ5 D3ZCG3 A0A2Y9L538 F7I689 U3DBM1 F7I1K7 G1S5J7 A0A2K6UP60 A0A2K6UP77 A0A2R9BKE4 K7DBP6 K7AIA9 A0A2R9BC50 A0A2R9BC32 A0A2J8KVP8 A0A2K6UPA6 K7D708 A0A2J8KVM7 G3SEK7 A0A2K5R5I0 A0A2K5R5H7 A0A2I2Y4X0 G3R8U3 H2RDJ5 A0A2J8UGJ8 A0A2K6CAX9 A0A2K6CAT7 A0A2K5X5Q0 A0A2K5X5T7 A0A2K5YC96 A0A2R9BKI0 A0A2I3MHU8 A0A2I3MQZ0 A0A2K5L7G8 A0A2K5L7M0 A0A2I3S1G0 A0A2J8KVL7 Q7Z3U7 Q7Z3U7-5 A0A2K5X628 A0A0D9QWX1
A0A151IH52 F4WH85 A0A195F3L0 A0A195B9D3 E2BVY3 A0A195E5V5 A0A158NPD6 A0A088AHQ3 A0A2A3E4W7 A0A154P146 K7J2C6 A0A0J7L4K9 A0A232FB04 A0A1B6LWA5 A0A1B6KIN6 A0A1Y1M6D5 A0A1W4X2P8 A0A0C9RWJ8 D6W829 A0A310SFV2 A0A0L7RES7 E2AXI0 A0A1L8DV77 E0VBS7 U4TUS9 N6TSW3 A0A0M9ABM2 A0A2M4B960 A0A2M4B8J4 A0A1B6CWH9 A0A0K8UL09 W8C084 A0A034V2P6 A0A1A9ZEU2 A0A0T6AXK1 A0A1A9XUY6 A0A1B0ASB9 A0A0N7ZAU2 A0A1B0FP44 A0A1A9UW08 A0A3R7NKU3 A0A0M4EPF9 M3YZ50 W5P2N3 A0A2Y9KYG8 U3DXG9 A0A2Y9L534 U6DIP7 A0A1S2ZBS3 A0A1S2ZBQ5 D3ZCG3 A0A2Y9L538 F7I689 U3DBM1 F7I1K7 G1S5J7 A0A2K6UP60 A0A2K6UP77 A0A2R9BKE4 K7DBP6 K7AIA9 A0A2R9BC50 A0A2R9BC32 A0A2J8KVP8 A0A2K6UPA6 K7D708 A0A2J8KVM7 G3SEK7 A0A2K5R5I0 A0A2K5R5H7 A0A2I2Y4X0 G3R8U3 H2RDJ5 A0A2J8UGJ8 A0A2K6CAX9 A0A2K6CAT7 A0A2K5X5Q0 A0A2K5X5T7 A0A2K5YC96 A0A2R9BKI0 A0A2I3MHU8 A0A2I3MQZ0 A0A2K5L7G8 A0A2K5L7M0 A0A2I3S1G0 A0A2J8KVL7 Q7Z3U7 Q7Z3U7-5 A0A2K5X628 A0A0D9QWX1
Pubmed
22118469
24508170
30249741
21719571
20798317
21347285
+ More
20075255 28648823 28004739 18362917 19820115 20566863 23537049 24495485 25348373 20809919 25243066 15057822 22673903 22722832 16136131 22398555 10470851 12168954 14702039 17974005 16541075 15489334 16219684 16301316 18691976 18669648 20068231 21269460 22814378 24275569
20075255 28648823 28004739 18362917 19820115 20566863 23537049 24495485 25348373 20809919 25243066 15057822 22673903 22722832 16136131 22398555 10470851 12168954 14702039 17974005 16541075 15489334 16219684 16301316 18691976 18669648 20068231 21269460 22814378 24275569
EMBL
RSAL01000085
RVE48352.1
AGBW02014365
OWR41783.1
NEVH01002541
PNF42354.1
+ More
KK107139 EZA57654.1 QOIP01000003 RLU24976.1 KQ982482 KYQ55860.1 KQ977643 KYN00858.1 GL888153 EGI66451.1 KQ981836 KYN35043.1 KQ976542 KYM81148.1 GL451079 EFN80131.1 KQ979608 KYN20267.1 ADTU01022393 KZ288371 PBC26735.1 KQ434796 KZC05646.1 AAZX01001570 LBMM01000654 KMQ97867.1 NNAY01000566 OXU27628.1 GEBQ01012014 JAT27963.1 GEBQ01028659 JAT11318.1 GEZM01042424 GEZM01042423 JAV79865.1 GBYB01013395 JAG83162.1 KQ971307 EFA11008.1 KQ768478 OAD53119.1 KQ414608 KOC69457.1 GL443583 EFN61865.1 GFDF01003740 JAV10344.1 DS235039 EEB10833.1 KB631602 ERL84532.1 APGK01020363 KB740193 ENN81133.1 KQ435689 KOX81217.1 GGFJ01000227 MBW49368.1 GGFJ01000228 MBW49369.1 GEDC01026625 GEDC01026554 GEDC01019436 JAS10673.1 JAS10744.1 JAS17862.1 GDHF01024930 JAI27384.1 GAMC01011556 JAB94999.1 GAKP01022188 JAC36764.1 LJIG01022567 KRT79887.1 JXJN01002712 GDRN01093911 GDRN01093910 JAI59800.1 CCAG010013125 QCYY01004474 ROT60957.1 CP012523 ALC38725.1 AEYP01066302 AEYP01066303 AEYP01066304 AEYP01066305 AEYP01066306 AEYP01066307 AEYP01066308 AEYP01066309 AEYP01066310 AEYP01066311 AMGL01081276 AMGL01081277 AMGL01081278 AMGL01081279 GAMT01006780 GAMS01005095 JAB05081.1 JAB18041.1 HAAF01010615 CCP82439.1 AABR07057348 GAMR01004380 GAMP01000503 JAB29552.1 JAB52252.1 ADFV01002067 ADFV01002068 ADFV01002069 ADFV01002070 ADFV01002071 ADFV01002072 ADFV01002073 AJFE02087527 AJFE02087528 AJFE02087529 AJFE02087530 AACZ04013012 AACZ04013013 AACZ04013014 AACZ04013015 AACZ04013016 GABE01008959 JAA35780.1 GABC01001268 GABF01004404 GABD01005171 NBAG03000333 JAA10070.1 JAA17741.1 JAA27929.1 PNI39072.1 PNI39077.1 GABE01008960 JAA35779.1 PNI39074.1 CABD030083319 CABD030083320 CABD030083321 CABD030083322 NDHI03003457 PNJ44408.1 AQIA01011395 AQIA01011396 AQIA01011397 AHZZ02002360 AHZZ02002361 AHZZ02002362 AHZZ02002363 PNI39073.1 AB017814 AB028963 AK092646 AL834320 BX537415 AL833066 AC026115 AC079035 BC136621 BC141817 BC142710 BC151241 AQIB01023188 AQIB01023189 AQIB01023190
KK107139 EZA57654.1 QOIP01000003 RLU24976.1 KQ982482 KYQ55860.1 KQ977643 KYN00858.1 GL888153 EGI66451.1 KQ981836 KYN35043.1 KQ976542 KYM81148.1 GL451079 EFN80131.1 KQ979608 KYN20267.1 ADTU01022393 KZ288371 PBC26735.1 KQ434796 KZC05646.1 AAZX01001570 LBMM01000654 KMQ97867.1 NNAY01000566 OXU27628.1 GEBQ01012014 JAT27963.1 GEBQ01028659 JAT11318.1 GEZM01042424 GEZM01042423 JAV79865.1 GBYB01013395 JAG83162.1 KQ971307 EFA11008.1 KQ768478 OAD53119.1 KQ414608 KOC69457.1 GL443583 EFN61865.1 GFDF01003740 JAV10344.1 DS235039 EEB10833.1 KB631602 ERL84532.1 APGK01020363 KB740193 ENN81133.1 KQ435689 KOX81217.1 GGFJ01000227 MBW49368.1 GGFJ01000228 MBW49369.1 GEDC01026625 GEDC01026554 GEDC01019436 JAS10673.1 JAS10744.1 JAS17862.1 GDHF01024930 JAI27384.1 GAMC01011556 JAB94999.1 GAKP01022188 JAC36764.1 LJIG01022567 KRT79887.1 JXJN01002712 GDRN01093911 GDRN01093910 JAI59800.1 CCAG010013125 QCYY01004474 ROT60957.1 CP012523 ALC38725.1 AEYP01066302 AEYP01066303 AEYP01066304 AEYP01066305 AEYP01066306 AEYP01066307 AEYP01066308 AEYP01066309 AEYP01066310 AEYP01066311 AMGL01081276 AMGL01081277 AMGL01081278 AMGL01081279 GAMT01006780 GAMS01005095 JAB05081.1 JAB18041.1 HAAF01010615 CCP82439.1 AABR07057348 GAMR01004380 GAMP01000503 JAB29552.1 JAB52252.1 ADFV01002067 ADFV01002068 ADFV01002069 ADFV01002070 ADFV01002071 ADFV01002072 ADFV01002073 AJFE02087527 AJFE02087528 AJFE02087529 AJFE02087530 AACZ04013012 AACZ04013013 AACZ04013014 AACZ04013015 AACZ04013016 GABE01008959 JAA35780.1 GABC01001268 GABF01004404 GABD01005171 NBAG03000333 JAA10070.1 JAA17741.1 JAA27929.1 PNI39072.1 PNI39077.1 GABE01008960 JAA35779.1 PNI39074.1 CABD030083319 CABD030083320 CABD030083321 CABD030083322 NDHI03003457 PNJ44408.1 AQIA01011395 AQIA01011396 AQIA01011397 AHZZ02002360 AHZZ02002361 AHZZ02002362 AHZZ02002363 PNI39073.1 AB017814 AB028963 AK092646 AL834320 BX537415 AL833066 AC026115 AC079035 BC136621 BC141817 BC142710 BC151241 AQIB01023188 AQIB01023189 AQIB01023190
Proteomes
UP000283053
UP000007151
UP000235965
UP000053097
UP000279307
UP000075809
+ More
UP000078542 UP000007755 UP000078541 UP000078540 UP000008237 UP000078492 UP000005205 UP000005203 UP000242457 UP000076502 UP000002358 UP000036403 UP000215335 UP000192223 UP000007266 UP000053825 UP000000311 UP000009046 UP000030742 UP000019118 UP000053105 UP000092445 UP000092443 UP000092460 UP000092444 UP000078200 UP000283509 UP000092553 UP000000715 UP000002356 UP000248482 UP000079721 UP000002494 UP000008225 UP000001073 UP000233220 UP000240080 UP000002277 UP000001519 UP000233040 UP000233120 UP000233100 UP000233140 UP000028761 UP000233060 UP000005640 UP000029965
UP000078542 UP000007755 UP000078541 UP000078540 UP000008237 UP000078492 UP000005205 UP000005203 UP000242457 UP000076502 UP000002358 UP000036403 UP000215335 UP000192223 UP000007266 UP000053825 UP000000311 UP000009046 UP000030742 UP000019118 UP000053105 UP000092445 UP000092443 UP000092460 UP000092444 UP000078200 UP000283509 UP000092553 UP000000715 UP000002356 UP000248482 UP000079721 UP000002494 UP000008225 UP000001073 UP000233220 UP000240080 UP000002277 UP000001519 UP000233040 UP000233120 UP000233100 UP000233140 UP000028761 UP000233060 UP000005640 UP000029965
Interpro
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
A0A3S2NZ94
A0A212EJV0
A0A2J7RNE8
A0A026WRE2
A0A3L8DX75
A0A151X6D3
+ More
A0A151IH52 F4WH85 A0A195F3L0 A0A195B9D3 E2BVY3 A0A195E5V5 A0A158NPD6 A0A088AHQ3 A0A2A3E4W7 A0A154P146 K7J2C6 A0A0J7L4K9 A0A232FB04 A0A1B6LWA5 A0A1B6KIN6 A0A1Y1M6D5 A0A1W4X2P8 A0A0C9RWJ8 D6W829 A0A310SFV2 A0A0L7RES7 E2AXI0 A0A1L8DV77 E0VBS7 U4TUS9 N6TSW3 A0A0M9ABM2 A0A2M4B960 A0A2M4B8J4 A0A1B6CWH9 A0A0K8UL09 W8C084 A0A034V2P6 A0A1A9ZEU2 A0A0T6AXK1 A0A1A9XUY6 A0A1B0ASB9 A0A0N7ZAU2 A0A1B0FP44 A0A1A9UW08 A0A3R7NKU3 A0A0M4EPF9 M3YZ50 W5P2N3 A0A2Y9KYG8 U3DXG9 A0A2Y9L534 U6DIP7 A0A1S2ZBS3 A0A1S2ZBQ5 D3ZCG3 A0A2Y9L538 F7I689 U3DBM1 F7I1K7 G1S5J7 A0A2K6UP60 A0A2K6UP77 A0A2R9BKE4 K7DBP6 K7AIA9 A0A2R9BC50 A0A2R9BC32 A0A2J8KVP8 A0A2K6UPA6 K7D708 A0A2J8KVM7 G3SEK7 A0A2K5R5I0 A0A2K5R5H7 A0A2I2Y4X0 G3R8U3 H2RDJ5 A0A2J8UGJ8 A0A2K6CAX9 A0A2K6CAT7 A0A2K5X5Q0 A0A2K5X5T7 A0A2K5YC96 A0A2R9BKI0 A0A2I3MHU8 A0A2I3MQZ0 A0A2K5L7G8 A0A2K5L7M0 A0A2I3S1G0 A0A2J8KVL7 Q7Z3U7 Q7Z3U7-5 A0A2K5X628 A0A0D9QWX1
A0A151IH52 F4WH85 A0A195F3L0 A0A195B9D3 E2BVY3 A0A195E5V5 A0A158NPD6 A0A088AHQ3 A0A2A3E4W7 A0A154P146 K7J2C6 A0A0J7L4K9 A0A232FB04 A0A1B6LWA5 A0A1B6KIN6 A0A1Y1M6D5 A0A1W4X2P8 A0A0C9RWJ8 D6W829 A0A310SFV2 A0A0L7RES7 E2AXI0 A0A1L8DV77 E0VBS7 U4TUS9 N6TSW3 A0A0M9ABM2 A0A2M4B960 A0A2M4B8J4 A0A1B6CWH9 A0A0K8UL09 W8C084 A0A034V2P6 A0A1A9ZEU2 A0A0T6AXK1 A0A1A9XUY6 A0A1B0ASB9 A0A0N7ZAU2 A0A1B0FP44 A0A1A9UW08 A0A3R7NKU3 A0A0M4EPF9 M3YZ50 W5P2N3 A0A2Y9KYG8 U3DXG9 A0A2Y9L534 U6DIP7 A0A1S2ZBS3 A0A1S2ZBQ5 D3ZCG3 A0A2Y9L538 F7I689 U3DBM1 F7I1K7 G1S5J7 A0A2K6UP60 A0A2K6UP77 A0A2R9BKE4 K7DBP6 K7AIA9 A0A2R9BC50 A0A2R9BC32 A0A2J8KVP8 A0A2K6UPA6 K7D708 A0A2J8KVM7 G3SEK7 A0A2K5R5I0 A0A2K5R5H7 A0A2I2Y4X0 G3R8U3 H2RDJ5 A0A2J8UGJ8 A0A2K6CAX9 A0A2K6CAT7 A0A2K5X5Q0 A0A2K5X5T7 A0A2K5YC96 A0A2R9BKI0 A0A2I3MHU8 A0A2I3MQZ0 A0A2K5L7G8 A0A2K5L7M0 A0A2I3S1G0 A0A2J8KVL7 Q7Z3U7 Q7Z3U7-5 A0A2K5X628 A0A0D9QWX1
Ontologies
GO
PANTHER
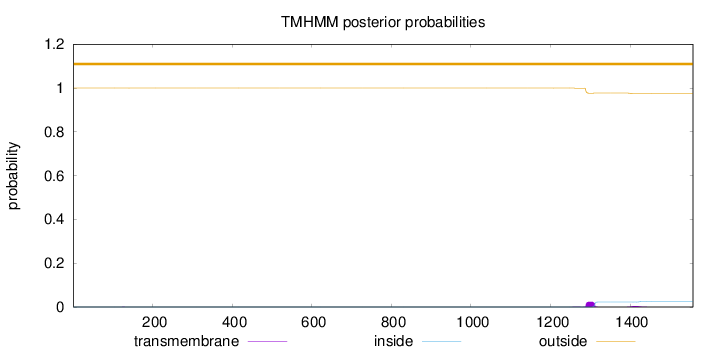
Topology
Length:
1558
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.634339999999998
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.00028
outside
1 - 1558
Population Genetic Test Statistics
Pi
26.648778
Theta
22.266233
Tajima's D
-1.437536
CLR
231.372091
CSRT
0.0692965351732413
Interpretation
Possibly Positive selection
