Gene
KWMTBOMO15901
Pre Gene Modal
BGIBMGA013403
Annotation
PREDICTED:_muscle_calcium_channel_subunit_alpha-1-like_isoform_X3_[Bombyx_mori]
Full name
Voltage-dependent L-type calcium channel subunit alpha
+ More
Voltage-dependent calcium channel type D subunit alpha-1
Muscle calcium channel subunit alpha-1
Voltage-dependent calcium channel type D subunit alpha-1
Muscle calcium channel subunit alpha-1
Alternative Name
MDL-alpha-1
Location in the cell
PlasmaMembrane Reliability : 4.375
Sequence
CDS
ATGGCATCGATAGAGCAAACCCCGACGGTGGCTACGACCCCGTACATGGAGAACGTCACTGACCCGACACTGGCAGCAGCCGCCGCCGCGAAGAAACAGGCTCGGGGCAGAGGAAAAGCGCAACCAGAAAAACCTAAGAGGAGCTTGATGTGCCTCACTCTGGACAACCCTTTACGGAAACTATGTTATAGCATCGTTGAGTGGAAACCATTTGAATACATGATCTTGACCACGATCTTCGCAAATTGCATCGCCTTAGCGGTATTCACACCGTACCCGTTCGGAGATACGAACAACACAAATCAAATACTGGAAAAAGTGGAATGGGTTTTCATGGTGATATTCACCGGGGAGTGCATCATCAAGATCATAGCGTACGGCTTCTTTTTTCATCCAGGAGCCTATCTTCGGAATACTTGGAACAGTCTAGATTTTACTATAGTCACAATCGGTATTGCGAGTATGATACAACAGTACCTTTCGATAGATTTGTTCGACGTAAAAGCACTGAGAGCTTTCAGGGTACTGAGGCCCTTACGACTCGTTTCCGGAGTACCTAGTCTTCAAATAGTTCTGAATTCCATCCTGAAAGCCATGGTGCCGCTCTTTCACATAGCCTTCCTTGTGCTGTTCGTGATCATCATCTATGCCATCATAGGCTTGGAGCTGTTCTCGGGAGTGCTCCATGAAACTTGCTTCAACAATGAAACAGGTGAGACAATGGCGAACCCTGCGCCTTGTAGTAGCACTCAAGGCGATGGTATGAAGTGCAAAGACAATATGACTTGCAAGGGTTATTGGGATGGACCCAATGATGGGATCACCAATTTTGACAACATCGGATACGCCATGTTGACTGTCTTCCAATGTATCACGCTTGAAGGCTGGACAGATATTATGTATGCGGTGGCCAACGTCAAAGGCAAAACGTGGGTTTGGATCTACTTCGTGACTTTGGTGATCTTCGGGAACTTCTTCGTCATGAATCTGATCCTGGGTGTGTTGTCAGGTGAGTTTTCCAAAGAAAGAGAGAAAGCGAAGAGCCGTGGCGATTTCCAGAAGTCACGCGAAAGGCAACAGCTGGAAGAAGACATGAAGGGGTATCTGAATTGGATCACAGTTGCGGAAGAGCTGGACTTGGAGAATGAACAGACGCAGAATGAAGATGATCAGGACGCCCAGGGCAAGAAAGATAGAAAGAGCAATGAAGAATCACAGAATAATTTGAATGTGAACGGGGATCAAGTACAAACATCAAGATCCTATAGATTCGATAAGGTCAATCGTCGATTACGTAGAGCCTGCAGAATAGCCGTGAAGACACAGACTTTCTATTGGGCAATCATCATATTGGTATTCCTCAATACGCTGGTGCTAGCTACTGAACATTACAATCAACCTCGATGGCTTGATTATTTCCAGAACTACGCCAACTATTTTTTCGTTGTTCTGTTCACCCTTGAGATGATGCTGAAGATGTACGCGCTAGGATTTCAGGGCTACTTCGTATCCCTATTCAATCGCTTCGATTGCTTCATCATGGTCTGCTCTGTGGTGGAATTGGCGCTTACTTTAACAGAGACCATACCGCAGCTCGGTATCTCCGTGCTGAGGTGTGTCAGGCTACTAAGAGTCTTCAAGGTTACCAAGTATTGGCGGTCTCTTTCTAATCTCGTAGCATCTCTGCTTAACTCCATCCAGTCCATCTTCTCCCTTCTTCTGCTGCTATTCTTGTTTATCGTGATCTTCGCTTTACTTGGGATGCAAGTGTTTGGTGGGAGATTCGACTACGCGCCGGAAGCTGAGAAAGAGAGGCATAACTTCGATTCTTTCTGGCAGGCAGTTCTGACCATGTTTCAGATACTGACAGGCGAAGATTGGAATGTAGTCATGTATGAGGGGATCAAGGCATACGGAGGCGCCGGGACTCCGGGCATGCTGGCGTCTATATACTTCATCGTTATATTCATTTGCGGCAATTATATTTTACTAAACGTATTCTTGGCCATCGCTGTCGACAACCTCGGCGACGCAGAGGAAATGGATGCAATCGAAAAACAGAAAGAGATGGCAGAAAACGCTGAAGCCGGTAATGTGGTTCTCGATGAAGAAAGAGGAGAAACTGATGATGAATATGTTAACGAGGAAGAAGGATATTCTAATGAAGGATCTGAGAATGAATGTGAAGAGACCGGTTCTGAAGAAGACGTGGAAGAACAGGAACAAGAGGAGAAAGAGAATCCTCAAGATATATTGGTCACTATTGACAAAAAACAAAATGGAGACGTTAACAAAAAAGAATCAGAGCCCCATGTAACAATTTCGGCCCGACCTCGAAGGTTATCGGATGTGAACATCAAAACGCAAGAGAAGCCTATACCTGATGCCAGTAGCTTCTTTATACTCTCCAAGGATAATTGTTTTCGGGTGGCCTGCTACAATATTCAAAAAAATTCCTGGTTCAAGAATCTTATTCTGATATGCATCTTGGCGTCTTCCGTCATGCTGGCTTTGGAGGACCCTGTTACCAGCAACTTGAACACCCCTACTGGGATAAATGAGGTTCTACGCAACGTGGATTACTTCTTCACGGCTGTTTTTACAATTGAGCTGCTATTGAAGCTCATTACATACGGTTTTATACTGCACAAAGATTCTTTCTGTCGTTCTGGATTCAACTTGCTCGATTTGCTGGTTGTGATCGTGTCTTTTATATCGTTGGACAGATCAAACAGCGTCAACTTCATTAAGATTTTGAGGGTCTTCCGCGTTTTAAGGCCGCTTCGTGCCATAAACAGAGCCAAAGGTCTGAAGCACGTAGTTCAATGCGTGATCGTTGCTATAAAGACAATCGGAAACATATTATTGGTGACAAATCTTCTACAATTCATGTTTGCCGTTATGGGAGTGCAAATGTTCAAGGGTAGATTTTTTAAATGTACAGATATTACGAAGGACAATGAAAATGATTGCCATGGCACGTATCTCATTTACGAGAACGGCGAGGCCATAACGAACGATCGTTTGTGGCTGCGCAACAAATTCAACTTCGACGACGTCCTCAACGGTATGCTGACTCTTTTCACCGTCTCCACGTACGAGGGATGGCCGGCTCTGCTCTCCACATCAATGGATTCTAATAAAGAGAATCACGGACCTATAGAAAATTCAAGGCCGTTCGTCGCGTTCTTCTATATCAGCTACATCATTGTCATTGCCTTTTTTATGGTCAATATCTTCGTGGGCTTTGTCATAGTCACGTTCCAGAAAGAAGGAGAGCAAGAGTTCAAGGACTGCGAGCTGGATAAGAACCAGAGGAACTGTATAGAGTTCGCACTAAAAGCCAAACCGATCAGAAGATACATTCCAAAGCATCGCATCCAGTACAAGACTTGGTGGTTTGTGACCTCGCAGCGTTTTGAAAACGTCATCTTTTTCTTCATCGTGCTAAACACCATCGCGCTGATGATGAAGTTCCATGGCTCCAAAACGGAATATAAAAAGGGACTTGACGTTCTTAATATGGGATTAACCACTGTCTTTACTTTGGAGTTCATCCTGAAATTGGCCGCATTTAGGTTTAAGAACTACTTCGGCGATGCTTGGAATACGACGGATTTCATTCTAGTCGTGGGGAGTATCATTGACATACTGGTGACGCAGGTCAACGAAAAAAAGGAGCTCGGAAGTTTCGATGCAAAAACTGAGAAAGGTCCCCTGTCAAAATACATTACGTTCTTTCGTCTCTTCCGGGCCATGAGGTTGATAAAACTCTTATCCAGAGGAGAGAGGATCAGGACTCTCCTGTGGACCTTCATCAAGTCATTCCAGGCTCTACCTTATGTCGCTTTATTGATTGTACTGCTATTCTTCATTTACGCTGTGGTTGGAATGCAGCTTTTCGGAAGAATTTACTTTGACGACGCAGTCATAACTGACCATAATAATTTCCAGACATTTCCGAACGCTCTCATGGTGCTCTTTCGGTCTGCCACAGGTGAAGCATGGCAGGAGATCATGATGGCACTGTCGCCCAATGAATACGAGCGTCCAGGTAATCCGCCTCACTGCATCAAAATCGACACAGACGGCCGCTACGTTGAAGATACAACAGAATGCGGTAGTTGGCTAGCGTTCCCCTACTTCATATCCTTCTCTTTGCTCTGTACATTCTTGATCATTAACTTATTTGTGGCGGTCATCATGGACAACTTTGATTATTTGACGAGAGATTGGTCCATTCTTGGCCCCCATCATCTGGACGAGTTTGTTCGGTTGTGGAGTGAATACGACCCCGACGCTAAAGGCAGGATCAAGCACTTAGACGTGGTGACGTTGCTGAGGAAAATAAGCCCGCCTTTAGGTTTCGGAAAGTTGTGCCCTCACAGGACTGCGTGCAAGCGTCTGGTGTCGATGAACATGCCTCTCAATTCCGACGGGACGGTCAATTTCAACGCGACCCTCTTTGCTGTTGTCCGAACTCAACTCCAAATAAAGACTACAGGTGTAATTGACGAATGCAATACCCAACTCCGCGCGATCATCAAAAGAGTTTGGAAGAGAACTTCTCCCAAGCTCCTGGACCAAGTGGTCCCGCCTCCCGGAGACCCGAACGAGATCACGGTGGGAAAATTCTACGCGACTTTCCTAATTCAAGATTACTTCCGGAGGTTCCGAAAACGAAAGGAGCAGGCTGCAGCCACCAACGTCCAGAAAACCTTCCAGGCTGGTATGAGGACTCTACACGAAGCCGGTCCAGAGCTGAAGAGAGCTATCTCCGGTAACCTAGATGAGATCGTCGAAATTGTGGAGCCTATGCACAGGCGCAACCACACATTATTCGGTGCTGTCCTAGCAAGTATCAAGAAACACGGTCGCGAGAGCTTGTATAGAAGAGAAAAACCACGTCAAGAAGTAAAAAGCTCTGCATCTGGCGATGGACCATCACTGGGCGCCATGATGCGACGGGAGCTTGGCTTGGATGGCAAAATACCTGAGGAAGAAGTGAAAGGTGACGAAGGTAACCACAGCGACGAAGAGATAGCGATGCAGCCTCTCCTTCACGGCAAAGATTCGGAAAGCATTTTCAAAGTTATCGAAAAAACATCCAACGATTTATTGCAGAGTATGCGAAACAACCGTCGTGACAAACAACGTGTACAAACCAACCACGTGCCTTACTGTGTAGAGAATCCTGCGGAGAAATTAATAATAAGGGTGCTGACCGAGCAAGGTCTAGGGAAGTACTGCGACAAAGACTTCATCGAGACCACGCTAAGAGAGATGCGAGAGGCGCTGGACCTAACACCGAACGAGATGGACACTGCTGCCAACCAAATAATAATGCAAGAGAGGATAATGAACGAAGCACAGCATCGACCGAGCGATGTGGAGAGTATTTTGTCAAGGCAGTACCATCCCTTCCACCAGCCTCCGATGCAGCCCCCTCCTAGGAAGAAATAA
Protein
MASIEQTPTVATTPYMENVTDPTLAAAAAAKKQARGRGKAQPEKPKRSLMCLTLDNPLRKLCYSIVEWKPFEYMILTTIFANCIALAVFTPYPFGDTNNTNQILEKVEWVFMVIFTGECIIKIIAYGFFFHPGAYLRNTWNSLDFTIVTIGIASMIQQYLSIDLFDVKALRAFRVLRPLRLVSGVPSLQIVLNSILKAMVPLFHIAFLVLFVIIIYAIIGLELFSGVLHETCFNNETGETMANPAPCSSTQGDGMKCKDNMTCKGYWDGPNDGITNFDNIGYAMLTVFQCITLEGWTDIMYAVANVKGKTWVWIYFVTLVIFGNFFVMNLILGVLSGEFSKEREKAKSRGDFQKSRERQQLEEDMKGYLNWITVAEELDLENEQTQNEDDQDAQGKKDRKSNEESQNNLNVNGDQVQTSRSYRFDKVNRRLRRACRIAVKTQTFYWAIIILVFLNTLVLATEHYNQPRWLDYFQNYANYFFVVLFTLEMMLKMYALGFQGYFVSLFNRFDCFIMVCSVVELALTLTETIPQLGISVLRCVRLLRVFKVTKYWRSLSNLVASLLNSIQSIFSLLLLLFLFIVIFALLGMQVFGGRFDYAPEAEKERHNFDSFWQAVLTMFQILTGEDWNVVMYEGIKAYGGAGTPGMLASIYFIVIFICGNYILLNVFLAIAVDNLGDAEEMDAIEKQKEMAENAEAGNVVLDEERGETDDEYVNEEEGYSNEGSENECEETGSEEDVEEQEQEEKENPQDILVTIDKKQNGDVNKKESEPHVTISARPRRLSDVNIKTQEKPIPDASSFFILSKDNCFRVACYNIQKNSWFKNLILICILASSVMLALEDPVTSNLNTPTGINEVLRNVDYFFTAVFTIELLLKLITYGFILHKDSFCRSGFNLLDLLVVIVSFISLDRSNSVNFIKILRVFRVLRPLRAINRAKGLKHVVQCVIVAIKTIGNILLVTNLLQFMFAVMGVQMFKGRFFKCTDITKDNENDCHGTYLIYENGEAITNDRLWLRNKFNFDDVLNGMLTLFTVSTYEGWPALLSTSMDSNKENHGPIENSRPFVAFFYISYIIVIAFFMVNIFVGFVIVTFQKEGEQEFKDCELDKNQRNCIEFALKAKPIRRYIPKHRIQYKTWWFVTSQRFENVIFFFIVLNTIALMMKFHGSKTEYKKGLDVLNMGLTTVFTLEFILKLAAFRFKNYFGDAWNTTDFILVVGSIIDILVTQVNEKKELGSFDAKTEKGPLSKYITFFRLFRAMRLIKLLSRGERIRTLLWTFIKSFQALPYVALLIVLLFFIYAVVGMQLFGRIYFDDAVITDHNNFQTFPNALMVLFRSATGEAWQEIMMALSPNEYERPGNPPHCIKIDTDGRYVEDTTECGSWLAFPYFISFSLLCTFLIINLFVAVIMDNFDYLTRDWSILGPHHLDEFVRLWSEYDPDAKGRIKHLDVVTLLRKISPPLGFGKLCPHRTACKRLVSMNMPLNSDGTVNFNATLFAVVRTQLQIKTTGVIDECNTQLRAIIKRVWKRTSPKLLDQVVPPPGDPNEITVGKFYATFLIQDYFRRFRKRKEQAAATNVQKTFQAGMRTLHEAGPELKRAISGNLDEIVEIVEPMHRRNHTLFGAVLASIKKHGRESLYRREKPRQEVKSSASGDGPSLGAMMRRELGLDGKIPEEEVKGDEGNHSDEEIAMQPLLHGKDSESIFKVIEKTSNDLLQSMRNNRRDKQRVQTNHVPYCVENPAEKLIIRVLTEQGLGKYCDKDFIETTLREMREALDLTPNEMDTAANQIIMQERIMNEAQHRPSDVESILSRQYHPFHQPPMQPPPRKK
Summary
Description
Voltage-sensitive calcium channels (VSCC) mediate the entry of calcium ions into excitable cells and are also involved in a variety of calcium-dependent processes, including muscle contraction, hormone or neurotransmitter release, gene expression, cell motility, cell division and cell death.
Voltage-sensitive calcium channels (VSCC) mediate the entry of calcium ions into excitable cells and are also involved in a variety of calcium-dependent processes, including muscle contraction, hormone or neurotransmitter release, gene expression, cell motility, cell division and cell death. Encodes a dihydropyridine- and diltiazem-sensitive current in larval body wall muscle. Vital for embryonic development.
Voltage-sensitive calcium channels (VSCC) mediate the entry of calcium ions into excitable cells and are also involved in a variety of calcium-dependent processes, including muscle contraction, hormone or neurotransmitter release, gene expression, cell motility, cell division and cell death (By similarity). MDL-alpha1 encodes a dihydropyridine- and diltiazem-sensitive current in larval body wall muscle.
Voltage-sensitive calcium channels (VSCC) mediate the entry of calcium ions into excitable cells and are also involved in a variety of calcium-dependent processes, including muscle contraction, hormone or neurotransmitter release, gene expression, cell motility, cell division and cell death. Encodes a dihydropyridine- and diltiazem-sensitive current in larval body wall muscle. Vital for embryonic development.
Voltage-sensitive calcium channels (VSCC) mediate the entry of calcium ions into excitable cells and are also involved in a variety of calcium-dependent processes, including muscle contraction, hormone or neurotransmitter release, gene expression, cell motility, cell division and cell death (By similarity). MDL-alpha1 encodes a dihydropyridine- and diltiazem-sensitive current in larval body wall muscle.
Similarity
Belongs to the calcium channel alpha-1 subunit (TC 1.A.1.11) family.
Keywords
Alternative splicing
Calcium
Calcium channel
Calcium transport
Complete proteome
Glycoprotein
Ion channel
Ion transport
Membrane
Metal-binding
Phosphoprotein
Reference proteome
Repeat
Transmembrane
Transmembrane helix
Transport
Voltage-gated channel
Feature
chain Voltage-dependent calcium channel type D subunit alpha-1
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A2H1V5W4
A0A2A4JH62
A0A0N1ICW1
H9JV40
A0A2A4JHK9
A0A194PPI5
+ More
A0A3S2TK45 A0A194PP78 A0A212F4Q3 A0A182J4Y3 A0A0J9R317 Q24270-3 A0A146LAM7 A0A0R1DSB5 M9PD04 A0A0R1DV93 A0A182N8Y2 A5YW88 A0A2H1WBU8 A0A1B6MRB3 A0A0Q9WRS5 A0A1W4VY04 A0A1B6L2C7 A0A1B6KE01 A0A1B6LPL5 A0A336KLA4 A0A1B6MJ45 A0A034V872 A0A084W5P2 A0A3B0K9Z1 A0A1A9W1K3 A0A0R1DN16 A0A2S1ZNL4 A0A0P6IY66 Q16QJ2 A0A1A9ZAA4 A0A1A9YAX7 A0A1W4VXA8 A0A0J9R2K5 A0A0J9R321 A0A182RAG6 A0A336MCG2 A0A336KKI5 A0A336MHW1 A0A336MC97 B3MKY4 A0A336MGI3 A0A1I8NY94 A0A1I8NYA2 A0A1I8NWX6 Q25452 A0A1A9UT84 A0A0A9X2A8 A0A139WPC9 A0A0A9WYS4 E0VY79 T1GVR3 A0A2S2Q3V7 A0A1W4X2P7 A0A0A9WWT5 A0A0A9WZR4 A0A2S1WM49 A0A0N5DJ56 A0A1W4X2N9 A0A1W4WRY0 A0A0J9R2K9 A0A1W4X2W4 A0A1W4WRY1 A0A0V1ME06 A0A182N8Y0 A0A0V1MD94 A0A0N5AZQ0 W5JR46 A0A0K0F0E8 A0A0N5BCI2 A0A0K0E6W3 A0A090MQP3 A0A2A2KHM2 A0A3Q2UF34 A0A2A2KHQ8 A0A0V1BDZ5 W5KJY4 A0A182MQB5 A0A0K8V6G5 A0A1B6J310 A0A1B6K0F5 A0A1B6IYS2 A0A1B6JJV1
A0A3S2TK45 A0A194PP78 A0A212F4Q3 A0A182J4Y3 A0A0J9R317 Q24270-3 A0A146LAM7 A0A0R1DSB5 M9PD04 A0A0R1DV93 A0A182N8Y2 A5YW88 A0A2H1WBU8 A0A1B6MRB3 A0A0Q9WRS5 A0A1W4VY04 A0A1B6L2C7 A0A1B6KE01 A0A1B6LPL5 A0A336KLA4 A0A1B6MJ45 A0A034V872 A0A084W5P2 A0A3B0K9Z1 A0A1A9W1K3 A0A0R1DN16 A0A2S1ZNL4 A0A0P6IY66 Q16QJ2 A0A1A9ZAA4 A0A1A9YAX7 A0A1W4VXA8 A0A0J9R2K5 A0A0J9R321 A0A182RAG6 A0A336MCG2 A0A336KKI5 A0A336MHW1 A0A336MC97 B3MKY4 A0A336MGI3 A0A1I8NY94 A0A1I8NYA2 A0A1I8NWX6 Q25452 A0A1A9UT84 A0A0A9X2A8 A0A139WPC9 A0A0A9WYS4 E0VY79 T1GVR3 A0A2S2Q3V7 A0A1W4X2P7 A0A0A9WWT5 A0A0A9WZR4 A0A2S1WM49 A0A0N5DJ56 A0A1W4X2N9 A0A1W4WRY0 A0A0J9R2K9 A0A1W4X2W4 A0A1W4WRY1 A0A0V1ME06 A0A182N8Y0 A0A0V1MD94 A0A0N5AZQ0 W5JR46 A0A0K0F0E8 A0A0N5BCI2 A0A0K0E6W3 A0A090MQP3 A0A2A2KHM2 A0A3Q2UF34 A0A2A2KHQ8 A0A0V1BDZ5 W5KJY4 A0A182MQB5 A0A0K8V6G5 A0A1B6J310 A0A1B6K0F5 A0A1B6IYS2 A0A1B6JJV1
Pubmed
EMBL
ODYU01000836
SOQ36207.1
NWSH01001536
PCG70924.1
LADJ01044508
KPJ21426.1
+ More
BABH01026241 BABH01026242 PCG70923.1 KQ459597 KPI95222.1 RSAL01000085 RVE48330.1 KPI95241.1 AGBW02010326 OWR48711.1 CM002910 KMY90461.1 U00690 AE014134 AY058268 GDHC01013994 JAQ04635.1 CM000157 KRJ98658.1 AGB93019.1 KRJ98662.1 EF595743 ABQ88369.1 ODYU01007597 SOQ50517.1 GEBQ01001497 JAT38480.1 CH963913 KRF98665.1 GEBQ01022182 JAT17795.1 GEBQ01030306 JAT09671.1 GEBQ01014334 JAT25643.1 UFQS01000565 UFQT01000565 SSX05001.1 SSX25363.1 GEBQ01004055 JAT35922.1 GAKP01019444 GAKP01019436 JAC39516.1 ATLV01020677 KE525304 KFB45536.1 OUUW01000006 SPP82466.1 KRJ98660.1 MF359245 AWK26948.1 GDUN01001113 JAN94806.1 CH477747 EAT36648.1 KMY90457.1 KMY90463.1 UFQT01000900 SSX27916.1 SSX04999.1 SSX25361.1 SSX27917.1 SSX27915.1 CH902620 EDV30642.2 SSX27913.1 Z31723 GBHO01030681 JAG12923.1 KQ971307 KYB29717.1 GBHO01030685 JAG12919.1 DS235844 EEB18335.1 CAQQ02090988 CAQQ02090989 CAQQ02090990 CAQQ02090991 GGMS01003201 MBY72404.1 GBHO01030682 JAG12922.1 GBHO01030683 JAG12921.1 MG973401 AWJ68254.1 KMY90462.1 JYDO01000127 KRZ69843.1 KRZ69841.1 ADMH02000580 ETN65778.1 LN609399 CEF60493.1 LIAE01008608 PAV73407.1 AADN05000356 AC171014 PAV73408.1 JYDH01000055 KRY35332.1 AXCM01000634 GDHF01017792 JAI34522.1 GECU01014145 JAS93561.1 GECU01002798 JAT04909.1 GECU01015631 JAS92075.1 GECU01008269 JAS99437.1
BABH01026241 BABH01026242 PCG70923.1 KQ459597 KPI95222.1 RSAL01000085 RVE48330.1 KPI95241.1 AGBW02010326 OWR48711.1 CM002910 KMY90461.1 U00690 AE014134 AY058268 GDHC01013994 JAQ04635.1 CM000157 KRJ98658.1 AGB93019.1 KRJ98662.1 EF595743 ABQ88369.1 ODYU01007597 SOQ50517.1 GEBQ01001497 JAT38480.1 CH963913 KRF98665.1 GEBQ01022182 JAT17795.1 GEBQ01030306 JAT09671.1 GEBQ01014334 JAT25643.1 UFQS01000565 UFQT01000565 SSX05001.1 SSX25363.1 GEBQ01004055 JAT35922.1 GAKP01019444 GAKP01019436 JAC39516.1 ATLV01020677 KE525304 KFB45536.1 OUUW01000006 SPP82466.1 KRJ98660.1 MF359245 AWK26948.1 GDUN01001113 JAN94806.1 CH477747 EAT36648.1 KMY90457.1 KMY90463.1 UFQT01000900 SSX27916.1 SSX04999.1 SSX25361.1 SSX27917.1 SSX27915.1 CH902620 EDV30642.2 SSX27913.1 Z31723 GBHO01030681 JAG12923.1 KQ971307 KYB29717.1 GBHO01030685 JAG12919.1 DS235844 EEB18335.1 CAQQ02090988 CAQQ02090989 CAQQ02090990 CAQQ02090991 GGMS01003201 MBY72404.1 GBHO01030682 JAG12922.1 GBHO01030683 JAG12921.1 MG973401 AWJ68254.1 KMY90462.1 JYDO01000127 KRZ69843.1 KRZ69841.1 ADMH02000580 ETN65778.1 LN609399 CEF60493.1 LIAE01008608 PAV73407.1 AADN05000356 AC171014 PAV73408.1 JYDH01000055 KRY35332.1 AXCM01000634 GDHF01017792 JAI34522.1 GECU01014145 JAS93561.1 GECU01002798 JAT04909.1 GECU01015631 JAS92075.1 GECU01008269 JAS99437.1
Proteomes
UP000218220
UP000053240
UP000005204
UP000053268
UP000283053
UP000007151
+ More
UP000075880 UP000000803 UP000002282 UP000075884 UP000007798 UP000192221 UP000030765 UP000268350 UP000091820 UP000008820 UP000092445 UP000092443 UP000075900 UP000007801 UP000095300 UP000095301 UP000078200 UP000007266 UP000009046 UP000015102 UP000192223 UP000046395 UP000054843 UP000046393 UP000000673 UP000035680 UP000046392 UP000035681 UP000218231 UP000000539 UP000054776 UP000018467 UP000075883
UP000075880 UP000000803 UP000002282 UP000075884 UP000007798 UP000192221 UP000030765 UP000268350 UP000091820 UP000008820 UP000092445 UP000092443 UP000075900 UP000007801 UP000095300 UP000095301 UP000078200 UP000007266 UP000009046 UP000015102 UP000192223 UP000046395 UP000054843 UP000046393 UP000000673 UP000035680 UP000046392 UP000035681 UP000218231 UP000000539 UP000054776 UP000018467 UP000075883
Interpro
SUPFAM
SSF53254
SSF53254
Gene 3D
ProteinModelPortal
A0A2H1V5W4
A0A2A4JH62
A0A0N1ICW1
H9JV40
A0A2A4JHK9
A0A194PPI5
+ More
A0A3S2TK45 A0A194PP78 A0A212F4Q3 A0A182J4Y3 A0A0J9R317 Q24270-3 A0A146LAM7 A0A0R1DSB5 M9PD04 A0A0R1DV93 A0A182N8Y2 A5YW88 A0A2H1WBU8 A0A1B6MRB3 A0A0Q9WRS5 A0A1W4VY04 A0A1B6L2C7 A0A1B6KE01 A0A1B6LPL5 A0A336KLA4 A0A1B6MJ45 A0A034V872 A0A084W5P2 A0A3B0K9Z1 A0A1A9W1K3 A0A0R1DN16 A0A2S1ZNL4 A0A0P6IY66 Q16QJ2 A0A1A9ZAA4 A0A1A9YAX7 A0A1W4VXA8 A0A0J9R2K5 A0A0J9R321 A0A182RAG6 A0A336MCG2 A0A336KKI5 A0A336MHW1 A0A336MC97 B3MKY4 A0A336MGI3 A0A1I8NY94 A0A1I8NYA2 A0A1I8NWX6 Q25452 A0A1A9UT84 A0A0A9X2A8 A0A139WPC9 A0A0A9WYS4 E0VY79 T1GVR3 A0A2S2Q3V7 A0A1W4X2P7 A0A0A9WWT5 A0A0A9WZR4 A0A2S1WM49 A0A0N5DJ56 A0A1W4X2N9 A0A1W4WRY0 A0A0J9R2K9 A0A1W4X2W4 A0A1W4WRY1 A0A0V1ME06 A0A182N8Y0 A0A0V1MD94 A0A0N5AZQ0 W5JR46 A0A0K0F0E8 A0A0N5BCI2 A0A0K0E6W3 A0A090MQP3 A0A2A2KHM2 A0A3Q2UF34 A0A2A2KHQ8 A0A0V1BDZ5 W5KJY4 A0A182MQB5 A0A0K8V6G5 A0A1B6J310 A0A1B6K0F5 A0A1B6IYS2 A0A1B6JJV1
A0A3S2TK45 A0A194PP78 A0A212F4Q3 A0A182J4Y3 A0A0J9R317 Q24270-3 A0A146LAM7 A0A0R1DSB5 M9PD04 A0A0R1DV93 A0A182N8Y2 A5YW88 A0A2H1WBU8 A0A1B6MRB3 A0A0Q9WRS5 A0A1W4VY04 A0A1B6L2C7 A0A1B6KE01 A0A1B6LPL5 A0A336KLA4 A0A1B6MJ45 A0A034V872 A0A084W5P2 A0A3B0K9Z1 A0A1A9W1K3 A0A0R1DN16 A0A2S1ZNL4 A0A0P6IY66 Q16QJ2 A0A1A9ZAA4 A0A1A9YAX7 A0A1W4VXA8 A0A0J9R2K5 A0A0J9R321 A0A182RAG6 A0A336MCG2 A0A336KKI5 A0A336MHW1 A0A336MC97 B3MKY4 A0A336MGI3 A0A1I8NY94 A0A1I8NYA2 A0A1I8NWX6 Q25452 A0A1A9UT84 A0A0A9X2A8 A0A139WPC9 A0A0A9WYS4 E0VY79 T1GVR3 A0A2S2Q3V7 A0A1W4X2P7 A0A0A9WWT5 A0A0A9WZR4 A0A2S1WM49 A0A0N5DJ56 A0A1W4X2N9 A0A1W4WRY0 A0A0J9R2K9 A0A1W4X2W4 A0A1W4WRY1 A0A0V1ME06 A0A182N8Y0 A0A0V1MD94 A0A0N5AZQ0 W5JR46 A0A0K0F0E8 A0A0N5BCI2 A0A0K0E6W3 A0A090MQP3 A0A2A2KHM2 A0A3Q2UF34 A0A2A2KHQ8 A0A0V1BDZ5 W5KJY4 A0A182MQB5 A0A0K8V6G5 A0A1B6J310 A0A1B6K0F5 A0A1B6IYS2 A0A1B6JJV1
PDB
5GJW
E-value=0,
Score=2559
Ontologies
GO
GO:0034765
GO:0005891
GO:0005245
GO:0016021
GO:0005244
GO:0005262
GO:0016322
GO:0042045
GO:0019722
GO:0006816
GO:0016324
GO:0070588
GO:0046872
GO:0006936
GO:0016323
GO:0070509
GO:0040017
GO:0045989
GO:0043051
GO:0035545
GO:0003301
GO:0048701
GO:0001947
GO:0008016
GO:0001822
GO:0005216
GO:0006811
GO:0016020
GO:0043565
GO:0003677
GO:0003707
GO:0006281
GO:0006259
GO:0055085
Topology
Subcellular location
Membrane
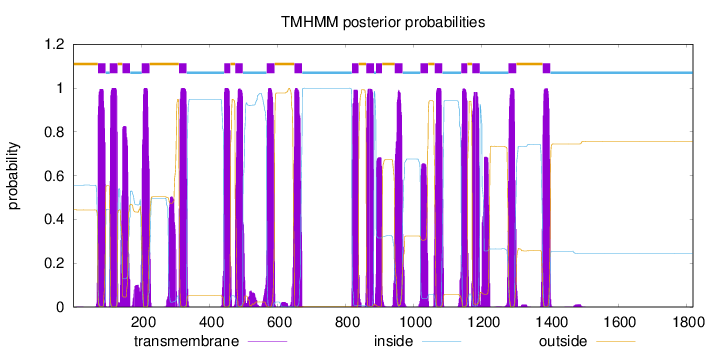
Length:
1820
Number of predicted TMHs:
19
Exp number of AAs in TMHs:
425.9204
Exp number, first 60 AAs:
0.01478
Total prob of N-in:
0.55610
outside
1 - 72
TMhelix
73 - 95
inside
96 - 107
TMhelix
108 - 130
outside
131 - 144
TMhelix
145 - 167
inside
168 - 201
TMhelix
202 - 224
outside
225 - 310
TMhelix
311 - 333
inside
334 - 443
TMhelix
444 - 461
outside
462 - 475
TMhelix
476 - 498
inside
499 - 568
TMhelix
569 - 591
outside
592 - 649
TMhelix
650 - 672
inside
673 - 818
TMhelix
819 - 838
outside
839 - 860
TMhelix
861 - 883
inside
884 - 889
TMhelix
890 - 907
outside
908 - 944
TMhelix
945 - 967
inside
968 - 1019
TMhelix
1020 - 1042
outside
1043 - 1061
TMhelix
1062 - 1084
inside
1085 - 1139
TMhelix
1140 - 1157
outside
1158 - 1171
TMhelix
1172 - 1194
inside
1195 - 1278
TMhelix
1279 - 1301
outside
1302 - 1378
TMhelix
1379 - 1401
inside
1402 - 1820
Population Genetic Test Statistics
Pi
264.861805
Theta
158.097867
Tajima's D
0.520532
CLR
0.707547
CSRT
0.524423778811059
Interpretation
Uncertain
