Gene
KWMTBOMO15898
Annotation
PREDICTED:_sporozoite_surface_protein_2-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.589
Sequence
CDS
ATGCGGATTTTGATACTGCTATCCATTTCCGCGGAGCTGGCGCTGACTGCTTCCATAAATGGACGATTATCCAACGATGACGATTTCAAGAGTTATGGAAAAAATGTGGAGCATCAAGAACCGGAAGAAAGAAGAGACTATAACATTCTAAGTGGATTTCCAGATAAGCCTAAAGAGGTAGCCCTTCCTTCAACGGTTGATCTCTTCAACGAATTTGAACCAAGCTTAACCGACAAACTCCATGACCTAACACCACTGAAACATAATCATCATAACGCTGCTACAGCGCCAGGAGTTCCAGATGAGTCTCAATTATCAGCACTGACCTTGATCCCGACAAAACATATCTTCCCGGATCAACCAGATCACCCCGTGAAATCACAACGTTCAAAAAAGCCTGATCATTCAGGGAAACCAGACAAACCTGGTCATCAAGAGAAACCAGACAAACCTGGTCATCAGGAGAAACCCAACAAGCCTGGTCATCACGAGAAACCGAACAAACCTAGTCATCAGGTGAAACCGGACAAACCTAGTCATCAGGAGAAACCGGACAAACCTGGTCATCAGGAGAAACCAAAAAAACCTGATCCTGATCATTTAGAAAAACCCAAGAAACCAGGTAATAATTACAAATCAGAGAAGTCTGATACTTCAGATAAACTAGAGAAGCCAGGCCACCACAAAGAAACAGAAAAATCAGAGAAATTGAGTCCTCTAGAGAAACTTGAGAAACCTGGTCATCACGAAAAACCAGATAAGCCTGATCATTTAGAGAAACTAGGCGCTCACGAAAAACCAGAGACGCCTGACCATTCAAAGAAACCAGGTCATCACGAAAAACCAGGGAAACTTGATCATTTAGAGAAACCAGGACATCACGAAAAACCAGGGAAACCTGATGATTCGGAAAAACCAAAGAAACCAGGACAACATGAAAAACCAGAGAAACCTGATCATTTAGAGAAACCAGAGAAACTAGGTCATCACGAAAAACCAGAAAAACCTCATTCAGAGAAACCAGAGAAGCCAGGCTATCACGGAAACCCAGATAAACCGTATCATCCAAAGAAACCAGGTGATCATGAAAAACCCAAGAAACCTGATCATTCAGAGAAACCAGAGAAACCTGATCATTCAGAGAAACCAGAGAAACCTAGTCATCACGAAAAACCAGAGAAACCTGGTGATGAAAAACCAGAGAAACCTGGTGATGAAAAACCAGAGAAACTTGGTGATGAAAAACCACAGGAACCTCATCATTCAGAAAAACCAGGGAAGCCAGGTCATAACGAAATACCAGGGAAACCAGGTGATGAAAAACCAGAGGAACTTGATCATTTAGAGAAACCAGAGAAACCTACTCATTCAGAGCAATCAGAGAAACCAGGAAATGAAAACCCAGAGAAACCCGATCATTCAGAGAAGCCAGAGAAACCTGGTGATGTAAACCCAGAAAAACCTGATCATTCAGAGAAACCAGAGGAACTTGAGAAGCCAGGTGATGAAAAACCAGAGAAACCAGGCGATCATGAAAAGCCAGAAAAACCTGATCCTTCAGAGAAACCAGAAAGCCCTGATAATTCAGCACATCCAAATTTGTCCGAAAATCCAGATAAACCAGAGAACGACGCCTACGGTTTACCTGGCCCAGAGAAACCAAACCATCCTTAA
Protein
MRILILLSISAELALTASINGRLSNDDDFKSYGKNVEHQEPEERRDYNILSGFPDKPKEVALPSTVDLFNEFEPSLTDKLHDLTPLKHNHHNAATAPGVPDESQLSALTLIPTKHIFPDQPDHPVKSQRSKKPDHSGKPDKPGHQEKPDKPGHQEKPNKPGHHEKPNKPSHQVKPDKPSHQEKPDKPGHQEKPKKPDPDHLEKPKKPGNNYKSEKSDTSDKLEKPGHHKETEKSEKLSPLEKLEKPGHHEKPDKPDHLEKLGAHEKPETPDHSKKPGHHEKPGKLDHLEKPGHHEKPGKPDDSEKPKKPGQHEKPEKPDHLEKPEKLGHHEKPEKPHSEKPEKPGYHGNPDKPYHPKKPGDHEKPKKPDHSEKPEKPDHSEKPEKPSHHEKPEKPGDEKPEKPGDEKPEKLGDEKPQEPHHSEKPGKPGHNEIPGKPGDEKPEELDHLEKPEKPTHSEQSEKPGNENPEKPDHSEKPEKPGDVNPEKPDHSEKPEELEKPGDEKPEKPGDHEKPEKPDPSEKPESPDNSAHPNLSENPDKPENDAYGLPGPEKPNHP
Summary
Feature
propeptide Removed by sortase
Uniprot
A0A1D7YLD9
A0A1H5AG60
A0A2G7FBW8
A0A3Q1JJ73
A0A162LWQ3
A0A2G5DNC1
+ More
A0A3Q1IB80 A0A3Q1I3G0 A0A3Q1JHV6 A0A2G5DN99 A0A2G5DNA7 A0A2G5DN97 A0A2G5DND8 A0A2G5DNA4 A0A3Q1JJB8 A0A0K0QP09 A0A3Q1I254 A0A3Q1JIZ0 A0A2G5DN88 A0A397MD90 A0A3D9J0B7 A0A2G5DNB7 A0A2G5DNC4 A0A2G5DNA1 A0A2G5DNB2 A0A2J7QN61 A0A0N0U4M6 A0A2G5DND0 A0A2G5DP16 A0A1S4DJW0 A0A1S4DJL2 A0A1S4DJK7 A0A1S4DJI6 A0A1S4DJL8 A0A1S4DJV0 A0A1S4DJK2 A0A1S4DJI1 A0A2U9C3E2 A0A1S4DJI5 A0A1S4DJM4 A0A1S4DJV5 A0A2G5DNB9 A0A2G5DNA8 A0A195D738 A0A139QNU6 F9LVS0 A0A1U7VRP7 A0A0T6B0P7
A0A3Q1IB80 A0A3Q1I3G0 A0A3Q1JHV6 A0A2G5DN99 A0A2G5DNA7 A0A2G5DN97 A0A2G5DND8 A0A2G5DNA4 A0A3Q1JJB8 A0A0K0QP09 A0A3Q1I254 A0A3Q1JIZ0 A0A2G5DN88 A0A397MD90 A0A3D9J0B7 A0A2G5DNB7 A0A2G5DNC4 A0A2G5DNA1 A0A2G5DNB2 A0A2J7QN61 A0A0N0U4M6 A0A2G5DND0 A0A2G5DP16 A0A1S4DJW0 A0A1S4DJL2 A0A1S4DJK7 A0A1S4DJI6 A0A1S4DJL8 A0A1S4DJV0 A0A1S4DJK2 A0A1S4DJI1 A0A2U9C3E2 A0A1S4DJI5 A0A1S4DJM4 A0A1S4DJV5 A0A2G5DNB9 A0A2G5DNA8 A0A195D738 A0A139QNU6 F9LVS0 A0A1U7VRP7 A0A0T6B0P7
EMBL
CP017248
AOR36413.1
FNTN01000001
SED41232.1
PEFT01000001
PIG78118.1
+ More
AZHC01000007 OAA46080.1 KZ305034 PIA44993.1 PIA45000.1 PIA45001.1 PIA44991.1 PIA44986.1 PIA44995.1 CP012103 AKR13228.1 PIA44990.1 QXCZ01000001 RIA22976.1 QREB01000001 RED66726.1 PIA44998.1 PIA44999.1 PIA44992.1 PIA45002.1 NEVH01013199 PNF30024.1 KQ435827 KOX71924.1 PIA45003.1 PIA44987.1 CP026254 AWP10603.1 PIA44989.1 PIA44988.1 KQ976750 KYN08705.1 LQZB01000149 KXU04230.1 AFUB01000019 EGU68650.1 LJIG01016296 KRT81027.1
AZHC01000007 OAA46080.1 KZ305034 PIA44993.1 PIA45000.1 PIA45001.1 PIA44991.1 PIA44986.1 PIA44995.1 CP012103 AKR13228.1 PIA44990.1 QXCZ01000001 RIA22976.1 QREB01000001 RED66726.1 PIA44998.1 PIA44999.1 PIA44992.1 PIA45002.1 NEVH01013199 PNF30024.1 KQ435827 KOX71924.1 PIA45003.1 PIA44987.1 CP026254 AWP10603.1 PIA44989.1 PIA44988.1 KQ976750 KYN08705.1 LQZB01000149 KXU04230.1 AFUB01000019 EGU68650.1 LJIG01016296 KRT81027.1
Proteomes
Pfam
Interpro
IPR027856
DUF4573
+ More
IPR008160 Collagen
IPR006792 Vicilin_N
IPR014710 RmlC-like_jellyroll
IPR011051 RmlC_Cupin_sf
IPR006045 Cupin_1
IPR000618 Insect_cuticle
IPR019931 LPXTG_anchor
IPR019948 Gram-positive_anchor
IPR004124 Glyco_hydro_33_N
IPR026856 Sialidase_fam
IPR011040 Sialidase
IPR005877 YSIRK_signal_dom
IPR036278 Sialidase_sf
IPR013320 ConA-like_dom_sf
IPR017853 Glycoside_hydrolase_SF
IPR000421 FA58C
IPR011098 G5_dom
IPR009003 Peptidase_S1_PA
IPR000933 Glyco_hydro_29
IPR003644 Calx_beta
IPR008979 Galactose-bd-like_sf
IPR038081 CalX-like_sf
IPR033336 SAXO1/2
IPR008160 Collagen
IPR006792 Vicilin_N
IPR014710 RmlC-like_jellyroll
IPR011051 RmlC_Cupin_sf
IPR006045 Cupin_1
IPR000618 Insect_cuticle
IPR019931 LPXTG_anchor
IPR019948 Gram-positive_anchor
IPR004124 Glyco_hydro_33_N
IPR026856 Sialidase_fam
IPR011040 Sialidase
IPR005877 YSIRK_signal_dom
IPR036278 Sialidase_sf
IPR013320 ConA-like_dom_sf
IPR017853 Glycoside_hydrolase_SF
IPR000421 FA58C
IPR011098 G5_dom
IPR009003 Peptidase_S1_PA
IPR000933 Glyco_hydro_29
IPR003644 Calx_beta
IPR008979 Galactose-bd-like_sf
IPR038081 CalX-like_sf
IPR033336 SAXO1/2
SUPFAM
Gene 3D
ProteinModelPortal
A0A1D7YLD9
A0A1H5AG60
A0A2G7FBW8
A0A3Q1JJ73
A0A162LWQ3
A0A2G5DNC1
+ More
A0A3Q1IB80 A0A3Q1I3G0 A0A3Q1JHV6 A0A2G5DN99 A0A2G5DNA7 A0A2G5DN97 A0A2G5DND8 A0A2G5DNA4 A0A3Q1JJB8 A0A0K0QP09 A0A3Q1I254 A0A3Q1JIZ0 A0A2G5DN88 A0A397MD90 A0A3D9J0B7 A0A2G5DNB7 A0A2G5DNC4 A0A2G5DNA1 A0A2G5DNB2 A0A2J7QN61 A0A0N0U4M6 A0A2G5DND0 A0A2G5DP16 A0A1S4DJW0 A0A1S4DJL2 A0A1S4DJK7 A0A1S4DJI6 A0A1S4DJL8 A0A1S4DJV0 A0A1S4DJK2 A0A1S4DJI1 A0A2U9C3E2 A0A1S4DJI5 A0A1S4DJM4 A0A1S4DJV5 A0A2G5DNB9 A0A2G5DNA8 A0A195D738 A0A139QNU6 F9LVS0 A0A1U7VRP7 A0A0T6B0P7
A0A3Q1IB80 A0A3Q1I3G0 A0A3Q1JHV6 A0A2G5DN99 A0A2G5DNA7 A0A2G5DN97 A0A2G5DND8 A0A2G5DNA4 A0A3Q1JJB8 A0A0K0QP09 A0A3Q1I254 A0A3Q1JIZ0 A0A2G5DN88 A0A397MD90 A0A3D9J0B7 A0A2G5DNB7 A0A2G5DNC4 A0A2G5DNA1 A0A2G5DNB2 A0A2J7QN61 A0A0N0U4M6 A0A2G5DND0 A0A2G5DP16 A0A1S4DJW0 A0A1S4DJL2 A0A1S4DJK7 A0A1S4DJI6 A0A1S4DJL8 A0A1S4DJV0 A0A1S4DJK2 A0A1S4DJI1 A0A2U9C3E2 A0A1S4DJI5 A0A1S4DJM4 A0A1S4DJV5 A0A2G5DNB9 A0A2G5DNA8 A0A195D738 A0A139QNU6 F9LVS0 A0A1U7VRP7 A0A0T6B0P7
Ontologies
GO
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 16,
Likelihood: 0.944929
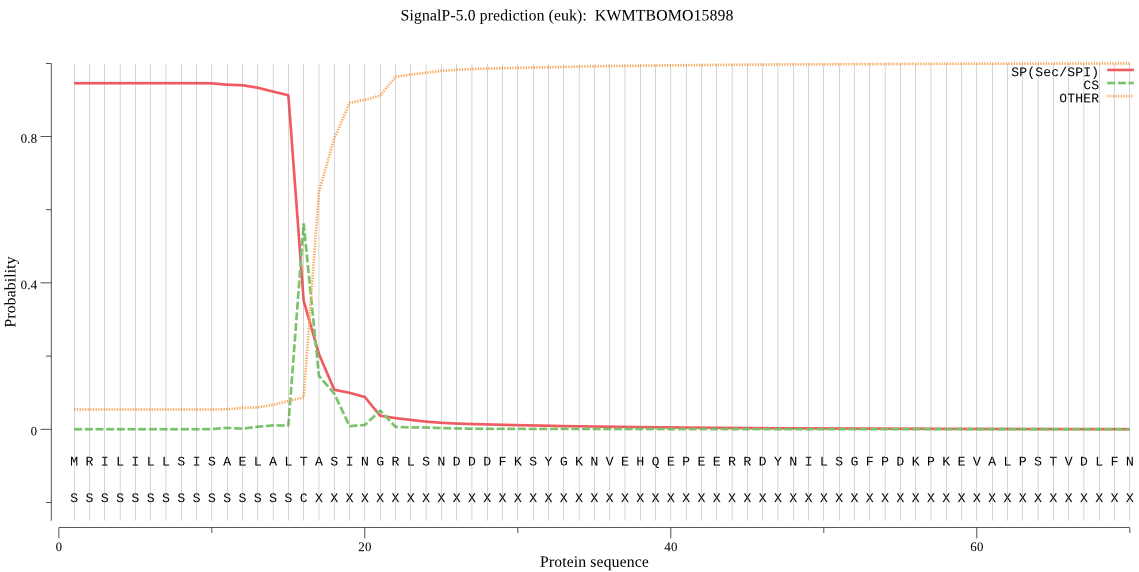
Length:
557
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03351
Exp number, first 60 AAs:
0.03351
Total prob of N-in:
0.00192
outside
1 - 557

Population Genetic Test Statistics
Pi
240.85668
Theta
174.880048
Tajima's D
0.727055
CLR
0.224269
CSRT
0.580270986450677
Interpretation
Uncertain
