Gene
KWMTBOMO15895
Annotation
PREDICTED:_dual_serine/threonine_and_tyrosine_protein_kinase-like_[Bombyx_mori]
Full name
Dual serine/threonine and tyrosine protein kinase
Alternative Name
Dusty protein kinase
Receptor-interacting serine/threonine-protein kinase 5
Receptor-interacting serine/threonine-protein kinase 5
Location in the cell
Cytoplasmic Reliability : 2.771
Sequence
CDS
ATGAGGATCAGCTGCGATATCGTCGACGGCATCCGCTACCTGCACTCGCTGGGTCTGGTGCACCGCGACATCAAGACGAAGAACGTCCTCCTGGACGGGGCGGACCGCGCGGCGCTGTCGGACCTCGGCTTCTGCACGGCCGGCGCGCTCATGTCGGGCTCCGTGGTGGGCACGCCGGTGCACATGGCGCCCGAGCTGCTGGCCGGGGACTACCACGCCTCCGTCGACGTGTACGCTTTCGGGATCTTGTTCTGGTACGTGTGCGCTGGTGACATAAAGCTACCGTCAGCTTTCGAAACTTTCCAAAACAAGGAACAGCTTTGGAGTAAGGTTAAAAGAGGTCTGAGACCTGAACGTCTACCTCAGTTCTCGGACGAGTGCTGGGAGATCATGGAGTCGTGCTGGGCCTCCGAGCCCTCCCAGCGGGCGCTCCTCGGGGACGTGCAGAACAAGCTGGAGAGGATCTTGGAGCAGGCTCTAAAGGACGAAAAGTCTCCTGGCTACCTCGCCCCGGACGAGCCAGACCTTAACGACGACTAG
Protein
MRISCDIVDGIRYLHSLGLVHRDIKTKNVLLDGADRAALSDLGFCTAGALMSGSVVGTPVHMAPELLAGDYHASVDVYAFGILFWYVCAGDIKLPSAFETFQNKEQLWSKVKRGLRPERLPQFSDECWEIMESCWASEPSQRALLGDVQNKLERILEQALKDEKSPGYLAPDEPDLNDD
Summary
Description
Acts as a positive regulator of ERK phosphorylation downstream of fibroblast growth factor-receptor activation. Involved in the regulation of both caspase-dependent apoptosis and caspase-independent cell death. In the skin, it plays a predominant role in suppressing caspase-dependent apoptosis in response to UV stress in a range of dermal cell types.
May act as a positive regulator of ERK phosphorylation downstream of fibroblast growth factor-receptor activation. May induce both caspase-dependent apoptosis and caspase-independent cell death (By similarity). Plays a role in the embryonic development.
May act as a positive regulator of ERK phosphorylation downstream of fibroblast growth factor-receptor activation. May induce both caspase-dependent apoptosis and caspase-independent cell death (By similarity). Plays a role in the embryonic development.
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family.
Keywords
ATP-binding
Cell junction
Cell membrane
Coiled coil
Complete proteome
Cytoplasm
Kinase
Membrane
Nucleotide-binding
Reference proteome
Transferase
Tyrosine-protein kinase
Alternative splicing
Developmental protein
Feature
chain Dual serine/threonine and tyrosine protein kinase
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A194PR80
A0A0N1IGG8
A0A2W1B383
A0A2H1W9T1
A0A2A4K4R5
A0A212FHW9
+ More
A0A0T6AY16 A0A0L7R6A4 N6U6V5 A0A139WP41 A0A151XGN8 A0A151ING0 A0A026W067 A0A151I231 E9G2L0 A0A195F8R4 E2A026 A0A158ND34 F4X0G5 W4XY32 E2C1A0 A0A224YX58 A0A224YLQ7 B7P7E5 L7LY21 A0A069DN16 A0A232F999 A0A224YNP0 A0A0N8E2F9 A0A131XNM5 A0A131YBQ2 A0A0C9RJV3 A0A0C9QGB0 A0A131YVD8 T1JTX5 A0A1A6HAT5 A0A0P5JCT6 A0A3M0JE54 A0A0M8ZSU1 A0A1E1X3G5 H1A4G1 A0A0J7KZX5 A0A3S3PT07 A0A1E1XRS9 A0A023GDF7 A0A060WIK8 A0A293LJW0 A0A2U3Z5F0 A0A0P6GFZ8 A0A3R7MGE3 A0A0P5LEW9 A0A0N8CEF3 A0A0P5TVG1 A0A0P5TIH0 A0A087ZMX2 A0A0P5SF87 A0A0P5M8C7 A0A0P5YJ63 A0A0P6ADU7 A0A0P5Y2E2 A0A0P6JU25 A0A0P5QFK0 A0A0P5T3J6 A0A2A3EDW5 A0A293M158 K7FSS8 A0A226DLP2 K7FST1 F1PEI7 A0A2D4H9J4 A0A287B254 F1S361 K7FSS3 A0A3Q0GST0 A0A1Y1K4A8 A0A1Y1K584 A0A3N0YGD4 A0A1Z5KVH0 A0A091HIL8 A0A2U3ZZM0 A0A3Q7RNP7 A0A067RB43 A0A2Y9TK65 A0A212CQJ6 A0A2R5LI47 A0A164XGZ9 A0A2Y9TKH7 A0A3L7HK17 Q4VSN4 A0A093ELT1 A0A384CTG9 Q4VSN1-2 A0A2P4T4C5 Q4VSN1 A0A091K862 V5HLW5 A0A131XSW8 A0A091NLQ3 A0A384A693 M7B5Q0
A0A0T6AY16 A0A0L7R6A4 N6U6V5 A0A139WP41 A0A151XGN8 A0A151ING0 A0A026W067 A0A151I231 E9G2L0 A0A195F8R4 E2A026 A0A158ND34 F4X0G5 W4XY32 E2C1A0 A0A224YX58 A0A224YLQ7 B7P7E5 L7LY21 A0A069DN16 A0A232F999 A0A224YNP0 A0A0N8E2F9 A0A131XNM5 A0A131YBQ2 A0A0C9RJV3 A0A0C9QGB0 A0A131YVD8 T1JTX5 A0A1A6HAT5 A0A0P5JCT6 A0A3M0JE54 A0A0M8ZSU1 A0A1E1X3G5 H1A4G1 A0A0J7KZX5 A0A3S3PT07 A0A1E1XRS9 A0A023GDF7 A0A060WIK8 A0A293LJW0 A0A2U3Z5F0 A0A0P6GFZ8 A0A3R7MGE3 A0A0P5LEW9 A0A0N8CEF3 A0A0P5TVG1 A0A0P5TIH0 A0A087ZMX2 A0A0P5SF87 A0A0P5M8C7 A0A0P5YJ63 A0A0P6ADU7 A0A0P5Y2E2 A0A0P6JU25 A0A0P5QFK0 A0A0P5T3J6 A0A2A3EDW5 A0A293M158 K7FSS8 A0A226DLP2 K7FST1 F1PEI7 A0A2D4H9J4 A0A287B254 F1S361 K7FSS3 A0A3Q0GST0 A0A1Y1K4A8 A0A1Y1K584 A0A3N0YGD4 A0A1Z5KVH0 A0A091HIL8 A0A2U3ZZM0 A0A3Q7RNP7 A0A067RB43 A0A2Y9TK65 A0A212CQJ6 A0A2R5LI47 A0A164XGZ9 A0A2Y9TKH7 A0A3L7HK17 Q4VSN4 A0A093ELT1 A0A384CTG9 Q4VSN1-2 A0A2P4T4C5 Q4VSN1 A0A091K862 V5HLW5 A0A131XSW8 A0A091NLQ3 A0A384A693 M7B5Q0
EC Number
2.7.12.1
Pubmed
EMBL
KQ459597
KPI95254.1
KQ460466
KPJ14600.1
KZ150391
PZC71049.1
+ More
ODYU01007222 SOQ49810.1 NWSH01000148 PCG79059.1 AGBW02008451 OWR53333.1 LJIG01022577 KRT79835.1 KQ414646 KOC66405.1 APGK01047273 KB741077 KB632432 ENN74312.1 ERL95551.1 KQ971307 KYB29627.1 KQ982173 KYQ59380.1 KQ976933 KYN07003.1 KK107609 QOIP01000014 EZA48519.1 RLU15104.1 KQ976547 KYM80927.1 GL732530 EFX86277.1 KQ981744 KYN36439.1 GL435543 EFN73209.1 ADTU01012291 GL888498 EGI59957.1 AAGJ04000566 GL451914 EFN78262.1 GFPF01007398 MAA18544.1 GFPF01007400 MAA18546.1 ABJB010666035 ABJB010750526 DS651463 EEC02517.1 GACK01009280 JAA55754.1 GBGP01000278 JAC84917.1 NNAY01000604 OXU27421.1 GFPF01007399 MAA18545.1 GDIQ01068396 JAN26341.1 GEFH01000756 JAP67825.1 GEDV01012599 JAP75958.1 GBYB01013562 JAG83329.1 GBYB01012356 GBYB01013563 JAG82123.1 JAG83330.1 GEDV01006107 JAP82450.1 CAEY01000486 LZPO01037266 OBS75401.1 GDIQ01201949 JAK49776.1 QRBI01000149 RMB99252.1 KQ435896 KOX69266.1 GFAC01005368 JAT93820.1 ABQF01074534 LBMM01001760 KMQ95799.1 NCKU01000345 RWS15880.1 GFAA01001450 JAU01985.1 GBBM01003531 JAC31887.1 FR904557 CDQ66806.1 GFWV01009038 MAA33767.1 GDIQ01033662 JAN61075.1 QCYY01004352 ROT61111.1 GDIQ01170615 JAK81110.1 GDIP01139823 JAL63891.1 GDIP01121422 JAL82292.1 GDIP01125789 JAL77925.1 GDIP01140579 JAL63135.1 GDIQ01183010 JAK68715.1 GDIP01058741 JAM44974.1 GDIP01030694 JAM73021.1 GDIP01064948 JAM38767.1 GDIQ01008359 JAN86378.1 GDIQ01117866 JAL33860.1 GDIP01133027 JAL70687.1 KZ288287 PBC29366.1 GFWV01009039 MAA33768.1 AGCU01153103 AGCU01153104 AGCU01153105 AGCU01153106 AGCU01153107 AGCU01153108 AGCU01153109 AGCU01153110 AGCU01153111 AGCU01153112 LNIX01000017 OXA45764.1 AAEX03018251 AAEX03018252 IACK01003771 LAA68647.1 AEMK02000070 GEZM01095810 JAV55331.1 GEZM01095811 JAV55330.1 RJVU01042601 ROL45207.1 GFJQ02007945 JAV99024.1 KL536230 KFO94515.1 KK852812 KDR15870.1 MKHE01000014 OWK08202.1 GGLE01005050 MBY09176.1 LRGB01000996 KZS14223.1 RAZU01000200 RLQ66289.1 AY641093 KK376193 KFV46062.1 AY208855 AY429681 AY641096 BC067573 BC118677 PPHD01009339 POI31198.1 KK546586 KFP32526.1 GANP01009925 JAB74543.1 GEFM01006636 JAP69160.1 KL385747 KFP89854.1 KB548129 EMP30840.1
ODYU01007222 SOQ49810.1 NWSH01000148 PCG79059.1 AGBW02008451 OWR53333.1 LJIG01022577 KRT79835.1 KQ414646 KOC66405.1 APGK01047273 KB741077 KB632432 ENN74312.1 ERL95551.1 KQ971307 KYB29627.1 KQ982173 KYQ59380.1 KQ976933 KYN07003.1 KK107609 QOIP01000014 EZA48519.1 RLU15104.1 KQ976547 KYM80927.1 GL732530 EFX86277.1 KQ981744 KYN36439.1 GL435543 EFN73209.1 ADTU01012291 GL888498 EGI59957.1 AAGJ04000566 GL451914 EFN78262.1 GFPF01007398 MAA18544.1 GFPF01007400 MAA18546.1 ABJB010666035 ABJB010750526 DS651463 EEC02517.1 GACK01009280 JAA55754.1 GBGP01000278 JAC84917.1 NNAY01000604 OXU27421.1 GFPF01007399 MAA18545.1 GDIQ01068396 JAN26341.1 GEFH01000756 JAP67825.1 GEDV01012599 JAP75958.1 GBYB01013562 JAG83329.1 GBYB01012356 GBYB01013563 JAG82123.1 JAG83330.1 GEDV01006107 JAP82450.1 CAEY01000486 LZPO01037266 OBS75401.1 GDIQ01201949 JAK49776.1 QRBI01000149 RMB99252.1 KQ435896 KOX69266.1 GFAC01005368 JAT93820.1 ABQF01074534 LBMM01001760 KMQ95799.1 NCKU01000345 RWS15880.1 GFAA01001450 JAU01985.1 GBBM01003531 JAC31887.1 FR904557 CDQ66806.1 GFWV01009038 MAA33767.1 GDIQ01033662 JAN61075.1 QCYY01004352 ROT61111.1 GDIQ01170615 JAK81110.1 GDIP01139823 JAL63891.1 GDIP01121422 JAL82292.1 GDIP01125789 JAL77925.1 GDIP01140579 JAL63135.1 GDIQ01183010 JAK68715.1 GDIP01058741 JAM44974.1 GDIP01030694 JAM73021.1 GDIP01064948 JAM38767.1 GDIQ01008359 JAN86378.1 GDIQ01117866 JAL33860.1 GDIP01133027 JAL70687.1 KZ288287 PBC29366.1 GFWV01009039 MAA33768.1 AGCU01153103 AGCU01153104 AGCU01153105 AGCU01153106 AGCU01153107 AGCU01153108 AGCU01153109 AGCU01153110 AGCU01153111 AGCU01153112 LNIX01000017 OXA45764.1 AAEX03018251 AAEX03018252 IACK01003771 LAA68647.1 AEMK02000070 GEZM01095810 JAV55331.1 GEZM01095811 JAV55330.1 RJVU01042601 ROL45207.1 GFJQ02007945 JAV99024.1 KL536230 KFO94515.1 KK852812 KDR15870.1 MKHE01000014 OWK08202.1 GGLE01005050 MBY09176.1 LRGB01000996 KZS14223.1 RAZU01000200 RLQ66289.1 AY641093 KK376193 KFV46062.1 AY208855 AY429681 AY641096 BC067573 BC118677 PPHD01009339 POI31198.1 KK546586 KFP32526.1 GANP01009925 JAB74543.1 GEFM01006636 JAP69160.1 KL385747 KFP89854.1 KB548129 EMP30840.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000007151
UP000053825
UP000019118
+ More
UP000030742 UP000007266 UP000075809 UP000078542 UP000053097 UP000279307 UP000078540 UP000000305 UP000078541 UP000000311 UP000005205 UP000007755 UP000007110 UP000008237 UP000001555 UP000215335 UP000015104 UP000092124 UP000269221 UP000053105 UP000007754 UP000036403 UP000285301 UP000193380 UP000245341 UP000283509 UP000005203 UP000242457 UP000007267 UP000198287 UP000002254 UP000008227 UP000189705 UP000245320 UP000286640 UP000027135 UP000248484 UP000076858 UP000273346 UP000261680 UP000000437 UP000261681 UP000031443
UP000030742 UP000007266 UP000075809 UP000078542 UP000053097 UP000279307 UP000078540 UP000000305 UP000078541 UP000000311 UP000005205 UP000007755 UP000007110 UP000008237 UP000001555 UP000215335 UP000015104 UP000092124 UP000269221 UP000053105 UP000007754 UP000036403 UP000285301 UP000193380 UP000245341 UP000283509 UP000005203 UP000242457 UP000007267 UP000198287 UP000002254 UP000008227 UP000189705 UP000245320 UP000286640 UP000027135 UP000248484 UP000076858 UP000273346 UP000261680 UP000000437 UP000261681 UP000031443
Interpro
Gene 3D
ProteinModelPortal
A0A194PR80
A0A0N1IGG8
A0A2W1B383
A0A2H1W9T1
A0A2A4K4R5
A0A212FHW9
+ More
A0A0T6AY16 A0A0L7R6A4 N6U6V5 A0A139WP41 A0A151XGN8 A0A151ING0 A0A026W067 A0A151I231 E9G2L0 A0A195F8R4 E2A026 A0A158ND34 F4X0G5 W4XY32 E2C1A0 A0A224YX58 A0A224YLQ7 B7P7E5 L7LY21 A0A069DN16 A0A232F999 A0A224YNP0 A0A0N8E2F9 A0A131XNM5 A0A131YBQ2 A0A0C9RJV3 A0A0C9QGB0 A0A131YVD8 T1JTX5 A0A1A6HAT5 A0A0P5JCT6 A0A3M0JE54 A0A0M8ZSU1 A0A1E1X3G5 H1A4G1 A0A0J7KZX5 A0A3S3PT07 A0A1E1XRS9 A0A023GDF7 A0A060WIK8 A0A293LJW0 A0A2U3Z5F0 A0A0P6GFZ8 A0A3R7MGE3 A0A0P5LEW9 A0A0N8CEF3 A0A0P5TVG1 A0A0P5TIH0 A0A087ZMX2 A0A0P5SF87 A0A0P5M8C7 A0A0P5YJ63 A0A0P6ADU7 A0A0P5Y2E2 A0A0P6JU25 A0A0P5QFK0 A0A0P5T3J6 A0A2A3EDW5 A0A293M158 K7FSS8 A0A226DLP2 K7FST1 F1PEI7 A0A2D4H9J4 A0A287B254 F1S361 K7FSS3 A0A3Q0GST0 A0A1Y1K4A8 A0A1Y1K584 A0A3N0YGD4 A0A1Z5KVH0 A0A091HIL8 A0A2U3ZZM0 A0A3Q7RNP7 A0A067RB43 A0A2Y9TK65 A0A212CQJ6 A0A2R5LI47 A0A164XGZ9 A0A2Y9TKH7 A0A3L7HK17 Q4VSN4 A0A093ELT1 A0A384CTG9 Q4VSN1-2 A0A2P4T4C5 Q4VSN1 A0A091K862 V5HLW5 A0A131XSW8 A0A091NLQ3 A0A384A693 M7B5Q0
A0A0T6AY16 A0A0L7R6A4 N6U6V5 A0A139WP41 A0A151XGN8 A0A151ING0 A0A026W067 A0A151I231 E9G2L0 A0A195F8R4 E2A026 A0A158ND34 F4X0G5 W4XY32 E2C1A0 A0A224YX58 A0A224YLQ7 B7P7E5 L7LY21 A0A069DN16 A0A232F999 A0A224YNP0 A0A0N8E2F9 A0A131XNM5 A0A131YBQ2 A0A0C9RJV3 A0A0C9QGB0 A0A131YVD8 T1JTX5 A0A1A6HAT5 A0A0P5JCT6 A0A3M0JE54 A0A0M8ZSU1 A0A1E1X3G5 H1A4G1 A0A0J7KZX5 A0A3S3PT07 A0A1E1XRS9 A0A023GDF7 A0A060WIK8 A0A293LJW0 A0A2U3Z5F0 A0A0P6GFZ8 A0A3R7MGE3 A0A0P5LEW9 A0A0N8CEF3 A0A0P5TVG1 A0A0P5TIH0 A0A087ZMX2 A0A0P5SF87 A0A0P5M8C7 A0A0P5YJ63 A0A0P6ADU7 A0A0P5Y2E2 A0A0P6JU25 A0A0P5QFK0 A0A0P5T3J6 A0A2A3EDW5 A0A293M158 K7FSS8 A0A226DLP2 K7FST1 F1PEI7 A0A2D4H9J4 A0A287B254 F1S361 K7FSS3 A0A3Q0GST0 A0A1Y1K4A8 A0A1Y1K584 A0A3N0YGD4 A0A1Z5KVH0 A0A091HIL8 A0A2U3ZZM0 A0A3Q7RNP7 A0A067RB43 A0A2Y9TK65 A0A212CQJ6 A0A2R5LI47 A0A164XGZ9 A0A2Y9TKH7 A0A3L7HK17 Q4VSN4 A0A093ELT1 A0A384CTG9 Q4VSN1-2 A0A2P4T4C5 Q4VSN1 A0A091K862 V5HLW5 A0A131XSW8 A0A091NLQ3 A0A384A693 M7B5Q0
PDB
5O26
E-value=1.57968e-15,
Score=197
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Detected in basolateral and apical membranes of all tubular epithelia. Detected at apical cell-cell junctions. Colocalized with FGF receptors to the cell membrane. With evidence from 1 publications.
Cell membrane Detected in basolateral and apical membranes of all tubular epithelia. Detected at apical cell-cell junctions. Colocalized with FGF receptors to the cell membrane. With evidence from 1 publications.
Apical cell membrane Detected in basolateral and apical membranes of all tubular epithelia. Detected at apical cell-cell junctions. Colocalized with FGF receptors to the cell membrane. With evidence from 1 publications.
Basolateral cell membrane Detected in basolateral and apical membranes of all tubular epithelia. Detected at apical cell-cell junctions. Colocalized with FGF receptors to the cell membrane. With evidence from 1 publications.
Cell junction Detected in basolateral and apical membranes of all tubular epithelia. Detected at apical cell-cell junctions. Colocalized with FGF receptors to the cell membrane. With evidence from 1 publications.
Cell membrane Detected in basolateral and apical membranes of all tubular epithelia. Detected at apical cell-cell junctions. Colocalized with FGF receptors to the cell membrane. With evidence from 1 publications.
Apical cell membrane Detected in basolateral and apical membranes of all tubular epithelia. Detected at apical cell-cell junctions. Colocalized with FGF receptors to the cell membrane. With evidence from 1 publications.
Basolateral cell membrane Detected in basolateral and apical membranes of all tubular epithelia. Detected at apical cell-cell junctions. Colocalized with FGF receptors to the cell membrane. With evidence from 1 publications.
Cell junction Detected in basolateral and apical membranes of all tubular epithelia. Detected at apical cell-cell junctions. Colocalized with FGF receptors to the cell membrane. With evidence from 1 publications.
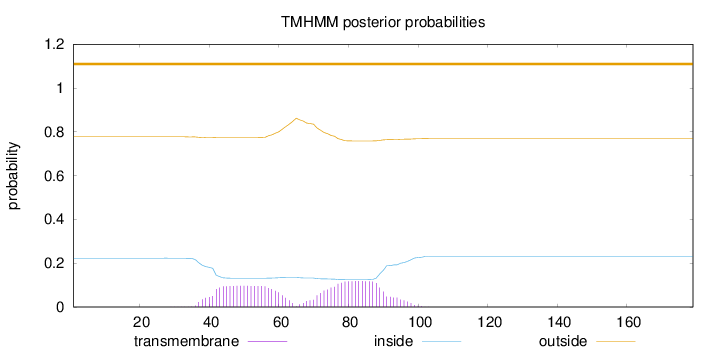
Length:
179
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.48827
Exp number, first 60 AAs:
1.92463
Total prob of N-in:
0.22300
outside
1 - 179
Population Genetic Test Statistics
Pi
43.857266
Theta
43.011215
Tajima's D
-0.431621
CLR
0.000008
CSRT
0.260886955652217
Interpretation
Uncertain
