Gene
KWMTBOMO15880
Pre Gene Modal
BGIBMGA013370
Annotation
PREDICTED:_talin-2-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.905
Sequence
CDS
ATGGAAGACGAGAAAGTCGAGAATAAGCCACGGTCTACCGGGACGCTGACTGCGAAGACACTTCAGTCTGGTCGCGACGCGCAGTTTGAGGAACTCAGCAAGAAGCTCCACACTGATGATAATGTCAAATGGCTCGACCACCACAAGACGCTTCGGGAGCTGTCCTTGGACGAGACCACGCCGTTCCTGCTGAAGAGGAAACTCTTCTACTCTGACAAGAACGTGGACGAAAGAGACCCGATGCAGCTCAATCTTCTCTACTTGCAAACCAGAGATGCAATACTCAACTTGACTCATCCGGTCACCGAAAAAGAAGGTGAGTTTTAA
Protein
MEDEKVENKPRSTGTLTAKTLQSGRDAQFEELSKKLHTDDNVKWLDHHKTLRELSLDETTPFLLKRKLFYSDKNVDERDPMQLNLLYLQTRDAILNLTHPVTEKEGEF
Summary
Uniprot
H9JV07
A0A1E1WHW2
A0A2W1BI91
A0A2H1VCC0
A0A1B6GVJ0
B7QM86
+ More
A0A1B6JKU4 A0A194PR75 V5GXZ8 A0A131XZA4 V5IGE5 A0A0N0PCT8 A0A1B6LJF3 A0A2A4JNE2 A0A1B6EM01 A0A1E1XFD7 A0A131YM95 A0A224Z8Y2 L7MJ63 L7MJY0 E9JDA6 A0A1W4XBC3 A0A131YGH8 A0A131XED9 A0A023GL65 A0A182SD41 H3C8Y1 A0A0S7L004 A0A2J7RK34 A0A1B6DNY2 A0A0P4W2W4 A0A212ETY4 A0A0P4W4Y4 A0A1B6BY27 A0A067R9F3 A0A0J7N8L8 A0A139WG31 A0A1Y1LTW5 A0A1Y1LRW5 A0A2P8Z921 A0A1Y1LQD1 A0A0S7KYX7 A0A2H1W5P8 A0A3P9P7K0 T1HTR5 A0A224XHD1 A0A069DY96 A0A2R5L7U9 H3BZC5 A0A1A8J134 S4RP70 A0A2R7W2L1 A0A0S7KYD2 A0A3B5QMV5 A0A0N0BGM5 A0A154PRL0 A0A158NZ32 W4VRP8 M3Z9X6 E9FVY2 A0A151WI83 Q6DEN1 A0A195BL13 A0A0N8P0V5 A0A0R3P6Z0 A0A158NZ33 Q4T143 Q568S4 A0A226EGS2 A1L1Z3 A0A3Q0IRT6 A0A3Q0ILZ7 A0A060Y5X6 Q3TKJ2 A0A2A3E5U7 A0A182X3R7 A0A182G5T2 A0A182I3A2 A0A182GR91 A0A182TPN4 Q7QJE3 Q176Z3 A0A182XYV8 A0A182RSM9 A0A3L8DYI0 A0A026WLW5 A0A0J9UH86 M9PF06 A0A182PQF6 A0A0R1DV89 A0A0Q5U2V2 A0A0N8ERZ1 W5MYM0 A0A182VF85 A0A3P8YN67 A0A195FSV9 F4WFN5 A0A2S2QV01 A0A1S3MAI4 A0A1S3MB54
A0A1B6JKU4 A0A194PR75 V5GXZ8 A0A131XZA4 V5IGE5 A0A0N0PCT8 A0A1B6LJF3 A0A2A4JNE2 A0A1B6EM01 A0A1E1XFD7 A0A131YM95 A0A224Z8Y2 L7MJ63 L7MJY0 E9JDA6 A0A1W4XBC3 A0A131YGH8 A0A131XED9 A0A023GL65 A0A182SD41 H3C8Y1 A0A0S7L004 A0A2J7RK34 A0A1B6DNY2 A0A0P4W2W4 A0A212ETY4 A0A0P4W4Y4 A0A1B6BY27 A0A067R9F3 A0A0J7N8L8 A0A139WG31 A0A1Y1LTW5 A0A1Y1LRW5 A0A2P8Z921 A0A1Y1LQD1 A0A0S7KYX7 A0A2H1W5P8 A0A3P9P7K0 T1HTR5 A0A224XHD1 A0A069DY96 A0A2R5L7U9 H3BZC5 A0A1A8J134 S4RP70 A0A2R7W2L1 A0A0S7KYD2 A0A3B5QMV5 A0A0N0BGM5 A0A154PRL0 A0A158NZ32 W4VRP8 M3Z9X6 E9FVY2 A0A151WI83 Q6DEN1 A0A195BL13 A0A0N8P0V5 A0A0R3P6Z0 A0A158NZ33 Q4T143 Q568S4 A0A226EGS2 A1L1Z3 A0A3Q0IRT6 A0A3Q0ILZ7 A0A060Y5X6 Q3TKJ2 A0A2A3E5U7 A0A182X3R7 A0A182G5T2 A0A182I3A2 A0A182GR91 A0A182TPN4 Q7QJE3 Q176Z3 A0A182XYV8 A0A182RSM9 A0A3L8DYI0 A0A026WLW5 A0A0J9UH86 M9PF06 A0A182PQF6 A0A0R1DV89 A0A0Q5U2V2 A0A0N8ERZ1 W5MYM0 A0A182VF85 A0A3P8YN67 A0A195FSV9 F4WFN5 A0A2S2QV01 A0A1S3MAI4 A0A1S3MB54
Pubmed
19121390
28756777
26354079
25765539
28503490
26830274
+ More
28797301 25576852 21282665 28049606 15496914 22118469 24845553 18362917 19820115 28004739 29403074 26334808 23542700 21347285 21292972 17994087 15632085 24755649 10349636 11042159 11076861 11217851 12466851 16141073 26483478 12364791 17510324 25244985 30249741 24508170 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26999592 25069045 21719571
28797301 25576852 21282665 28049606 15496914 22118469 24845553 18362917 19820115 28004739 29403074 26334808 23542700 21347285 21292972 17994087 15632085 24755649 10349636 11042159 11076861 11217851 12466851 16141073 26483478 12364791 17510324 25244985 30249741 24508170 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26999592 25069045 21719571
EMBL
BABH01037489
GDQN01004547
JAT86507.1
KZ150183
PZC72480.1
ODYU01001771
+ More
SOQ38436.1 GECZ01003328 JAS66441.1 ABJB010002632 ABJB010062123 ABJB010379358 ABJB010428970 ABJB010506374 ABJB010562744 ABJB010575441 ABJB010633491 ABJB010781916 ABJB010798695 DS969850 EEC19958.1 GECU01007852 JAS99854.1 KQ459597 KPI95249.1 GALX01003213 JAB65253.1 GEFM01003222 JAP72574.1 GANP01006827 JAB77641.1 KQ460466 KPJ14605.1 GEBQ01016146 JAT23831.1 NWSH01000974 PCG73339.1 GECZ01030847 JAS38922.1 GFAC01001233 JAT97955.1 GEDV01008164 JAP80393.1 GFPF01012237 MAA23383.1 GACK01001711 JAA63323.1 GACK01001466 JAA63568.1 GL771866 EFZ09348.1 GEDV01011536 JAP77021.1 GEFH01003634 JAP64947.1 GBBM01000771 JAC34647.1 GBYX01192628 GBYX01192624 GBYX01192623 GBYX01192620 JAO82251.1 NEVH01002981 PNF41181.1 GEDC01019901 GEDC01009902 GEDC01003946 JAS17397.1 JAS27396.1 JAS33352.1 GDRN01089357 JAI60682.1 AGBW02012523 OWR44939.1 GDRN01089358 JAI60681.1 GEDC01031369 GEDC01020851 GEDC01015209 JAS05929.1 JAS16447.1 JAS22089.1 KK852657 KDR19212.1 LBMM01008317 KMQ88985.1 KQ971351 KYB26811.1 GEZM01051359 GEZM01051357 JAV75016.1 GEZM01051360 GEZM01051358 JAV75010.1 PYGN01000142 PSN52989.1 GEZM01051361 JAV75008.1 GBYX01192625 GBYX01192621 JAO82249.1 ODYU01006474 SOQ48343.1 ACPB03000518 ACPB03000519 GFTR01008985 JAW07441.1 GBGD01000014 JAC88875.1 GGLE01001409 MBY05535.1 HAED01015993 SBR02438.1 KK854264 PTY13912.1 GBYX01192626 GBYX01192622 JAO82248.1 KQ435782 KOX74764.1 KQ435087 KZC14531.1 ADTU01004540 ADTU01004541 ADTU01004542 GANO01000813 JAB59058.1 GL732525 EFX88629.1 KQ983089 KYQ47547.1 BC077078 AAH77078.1 KQ976444 KYM86235.1 CH902618 KPU78180.1 CH379067 KRT07357.1 CAAE01010725 CAF93389.1 BC092742 AAH92742.1 LNIX01000004 OXA56480.1 BC129276 AAI29277.1 FR907704 CDQ87146.1 AK166970 BAE39153.1 KZ288358 PBC27080.1 JXUM01043900 KQ561380 KXJ78773.1 APCN01000204 JXUM01082049 JXUM01082050 JXUM01082051 KQ563286 KXJ74128.1 AAAB01008807 EAA04618.3 CH477380 EAT42216.1 QOIP01000003 RLU24868.1 KK107161 EZA56621.1 CM002912 KMY98410.1 AE014296 AGB94284.1 CM000159 KRK00985.1 CH954178 KQS43439.1 GDUN01000922 JAN94997.1 AHAT01013409 KQ981276 KYN43533.1 GL888120 EGI67012.1 GGMS01012351 MBY81554.1
SOQ38436.1 GECZ01003328 JAS66441.1 ABJB010002632 ABJB010062123 ABJB010379358 ABJB010428970 ABJB010506374 ABJB010562744 ABJB010575441 ABJB010633491 ABJB010781916 ABJB010798695 DS969850 EEC19958.1 GECU01007852 JAS99854.1 KQ459597 KPI95249.1 GALX01003213 JAB65253.1 GEFM01003222 JAP72574.1 GANP01006827 JAB77641.1 KQ460466 KPJ14605.1 GEBQ01016146 JAT23831.1 NWSH01000974 PCG73339.1 GECZ01030847 JAS38922.1 GFAC01001233 JAT97955.1 GEDV01008164 JAP80393.1 GFPF01012237 MAA23383.1 GACK01001711 JAA63323.1 GACK01001466 JAA63568.1 GL771866 EFZ09348.1 GEDV01011536 JAP77021.1 GEFH01003634 JAP64947.1 GBBM01000771 JAC34647.1 GBYX01192628 GBYX01192624 GBYX01192623 GBYX01192620 JAO82251.1 NEVH01002981 PNF41181.1 GEDC01019901 GEDC01009902 GEDC01003946 JAS17397.1 JAS27396.1 JAS33352.1 GDRN01089357 JAI60682.1 AGBW02012523 OWR44939.1 GDRN01089358 JAI60681.1 GEDC01031369 GEDC01020851 GEDC01015209 JAS05929.1 JAS16447.1 JAS22089.1 KK852657 KDR19212.1 LBMM01008317 KMQ88985.1 KQ971351 KYB26811.1 GEZM01051359 GEZM01051357 JAV75016.1 GEZM01051360 GEZM01051358 JAV75010.1 PYGN01000142 PSN52989.1 GEZM01051361 JAV75008.1 GBYX01192625 GBYX01192621 JAO82249.1 ODYU01006474 SOQ48343.1 ACPB03000518 ACPB03000519 GFTR01008985 JAW07441.1 GBGD01000014 JAC88875.1 GGLE01001409 MBY05535.1 HAED01015993 SBR02438.1 KK854264 PTY13912.1 GBYX01192626 GBYX01192622 JAO82248.1 KQ435782 KOX74764.1 KQ435087 KZC14531.1 ADTU01004540 ADTU01004541 ADTU01004542 GANO01000813 JAB59058.1 GL732525 EFX88629.1 KQ983089 KYQ47547.1 BC077078 AAH77078.1 KQ976444 KYM86235.1 CH902618 KPU78180.1 CH379067 KRT07357.1 CAAE01010725 CAF93389.1 BC092742 AAH92742.1 LNIX01000004 OXA56480.1 BC129276 AAI29277.1 FR907704 CDQ87146.1 AK166970 BAE39153.1 KZ288358 PBC27080.1 JXUM01043900 KQ561380 KXJ78773.1 APCN01000204 JXUM01082049 JXUM01082050 JXUM01082051 KQ563286 KXJ74128.1 AAAB01008807 EAA04618.3 CH477380 EAT42216.1 QOIP01000003 RLU24868.1 KK107161 EZA56621.1 CM002912 KMY98410.1 AE014296 AGB94284.1 CM000159 KRK00985.1 CH954178 KQS43439.1 GDUN01000922 JAN94997.1 AHAT01013409 KQ981276 KYN43533.1 GL888120 EGI67012.1 GGMS01012351 MBY81554.1
Proteomes
UP000005204
UP000001555
UP000053268
UP000053240
UP000218220
UP000192223
+ More
UP000075901 UP000007303 UP000235965 UP000007151 UP000027135 UP000036403 UP000007266 UP000245037 UP000242638 UP000015103 UP000245300 UP000002852 UP000053105 UP000076502 UP000005205 UP000001073 UP000000305 UP000075809 UP000078540 UP000007801 UP000001819 UP000198287 UP000079169 UP000193380 UP000242457 UP000076407 UP000069940 UP000249989 UP000075840 UP000075902 UP000007062 UP000008820 UP000076408 UP000075900 UP000279307 UP000053097 UP000000803 UP000075885 UP000002282 UP000008711 UP000018468 UP000075903 UP000265140 UP000078541 UP000007755 UP000087266
UP000075901 UP000007303 UP000235965 UP000007151 UP000027135 UP000036403 UP000007266 UP000245037 UP000242638 UP000015103 UP000245300 UP000002852 UP000053105 UP000076502 UP000005205 UP000001073 UP000000305 UP000075809 UP000078540 UP000007801 UP000001819 UP000198287 UP000079169 UP000193380 UP000242457 UP000076407 UP000069940 UP000249989 UP000075840 UP000075902 UP000007062 UP000008820 UP000076408 UP000075900 UP000279307 UP000053097 UP000000803 UP000075885 UP000002282 UP000008711 UP000018468 UP000075903 UP000265140 UP000078541 UP000007755 UP000087266
Pfam
Interpro
IPR018979
FERM_N
+ More
IPR030224 Sla2_fam
IPR029071 Ubiquitin-like_domsf
IPR000299 FERM_domain
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR032425 FERM_f0
IPR019748 FERM_central
IPR035963 FERM_2
IPR019749 Band_41_domain
IPR037843 Kindlin/fermitin
IPR006578 MADF-dom
IPR004210 BESS_motif
IPR019747 FERM_CS
IPR002558 ILWEQ_dom
IPR037438 Talin1/2-RS
IPR015009 Vinculin-bd_dom
IPR011993 PH-like_dom_sf
IPR036723 Alpha-catenin/vinculin-like_sf
IPR035964 I/LWEQ_dom_sf
IPR015224 Talin_cent
IPR036476 Talin_cent_sf
IPR002404 IRS_PTB
IPR006077 Vinculin/catenin
IPR030224 Sla2_fam
IPR029071 Ubiquitin-like_domsf
IPR000299 FERM_domain
IPR014352 FERM/acyl-CoA-bd_prot_sf
IPR032425 FERM_f0
IPR019748 FERM_central
IPR035963 FERM_2
IPR019749 Band_41_domain
IPR037843 Kindlin/fermitin
IPR006578 MADF-dom
IPR004210 BESS_motif
IPR019747 FERM_CS
IPR002558 ILWEQ_dom
IPR037438 Talin1/2-RS
IPR015009 Vinculin-bd_dom
IPR011993 PH-like_dom_sf
IPR036723 Alpha-catenin/vinculin-like_sf
IPR035964 I/LWEQ_dom_sf
IPR015224 Talin_cent
IPR036476 Talin_cent_sf
IPR002404 IRS_PTB
IPR006077 Vinculin/catenin
SUPFAM
Gene 3D
ProteinModelPortal
H9JV07
A0A1E1WHW2
A0A2W1BI91
A0A2H1VCC0
A0A1B6GVJ0
B7QM86
+ More
A0A1B6JKU4 A0A194PR75 V5GXZ8 A0A131XZA4 V5IGE5 A0A0N0PCT8 A0A1B6LJF3 A0A2A4JNE2 A0A1B6EM01 A0A1E1XFD7 A0A131YM95 A0A224Z8Y2 L7MJ63 L7MJY0 E9JDA6 A0A1W4XBC3 A0A131YGH8 A0A131XED9 A0A023GL65 A0A182SD41 H3C8Y1 A0A0S7L004 A0A2J7RK34 A0A1B6DNY2 A0A0P4W2W4 A0A212ETY4 A0A0P4W4Y4 A0A1B6BY27 A0A067R9F3 A0A0J7N8L8 A0A139WG31 A0A1Y1LTW5 A0A1Y1LRW5 A0A2P8Z921 A0A1Y1LQD1 A0A0S7KYX7 A0A2H1W5P8 A0A3P9P7K0 T1HTR5 A0A224XHD1 A0A069DY96 A0A2R5L7U9 H3BZC5 A0A1A8J134 S4RP70 A0A2R7W2L1 A0A0S7KYD2 A0A3B5QMV5 A0A0N0BGM5 A0A154PRL0 A0A158NZ32 W4VRP8 M3Z9X6 E9FVY2 A0A151WI83 Q6DEN1 A0A195BL13 A0A0N8P0V5 A0A0R3P6Z0 A0A158NZ33 Q4T143 Q568S4 A0A226EGS2 A1L1Z3 A0A3Q0IRT6 A0A3Q0ILZ7 A0A060Y5X6 Q3TKJ2 A0A2A3E5U7 A0A182X3R7 A0A182G5T2 A0A182I3A2 A0A182GR91 A0A182TPN4 Q7QJE3 Q176Z3 A0A182XYV8 A0A182RSM9 A0A3L8DYI0 A0A026WLW5 A0A0J9UH86 M9PF06 A0A182PQF6 A0A0R1DV89 A0A0Q5U2V2 A0A0N8ERZ1 W5MYM0 A0A182VF85 A0A3P8YN67 A0A195FSV9 F4WFN5 A0A2S2QV01 A0A1S3MAI4 A0A1S3MB54
A0A1B6JKU4 A0A194PR75 V5GXZ8 A0A131XZA4 V5IGE5 A0A0N0PCT8 A0A1B6LJF3 A0A2A4JNE2 A0A1B6EM01 A0A1E1XFD7 A0A131YM95 A0A224Z8Y2 L7MJ63 L7MJY0 E9JDA6 A0A1W4XBC3 A0A131YGH8 A0A131XED9 A0A023GL65 A0A182SD41 H3C8Y1 A0A0S7L004 A0A2J7RK34 A0A1B6DNY2 A0A0P4W2W4 A0A212ETY4 A0A0P4W4Y4 A0A1B6BY27 A0A067R9F3 A0A0J7N8L8 A0A139WG31 A0A1Y1LTW5 A0A1Y1LRW5 A0A2P8Z921 A0A1Y1LQD1 A0A0S7KYX7 A0A2H1W5P8 A0A3P9P7K0 T1HTR5 A0A224XHD1 A0A069DY96 A0A2R5L7U9 H3BZC5 A0A1A8J134 S4RP70 A0A2R7W2L1 A0A0S7KYD2 A0A3B5QMV5 A0A0N0BGM5 A0A154PRL0 A0A158NZ32 W4VRP8 M3Z9X6 E9FVY2 A0A151WI83 Q6DEN1 A0A195BL13 A0A0N8P0V5 A0A0R3P6Z0 A0A158NZ33 Q4T143 Q568S4 A0A226EGS2 A1L1Z3 A0A3Q0IRT6 A0A3Q0ILZ7 A0A060Y5X6 Q3TKJ2 A0A2A3E5U7 A0A182X3R7 A0A182G5T2 A0A182I3A2 A0A182GR91 A0A182TPN4 Q7QJE3 Q176Z3 A0A182XYV8 A0A182RSM9 A0A3L8DYI0 A0A026WLW5 A0A0J9UH86 M9PF06 A0A182PQF6 A0A0R1DV89 A0A0Q5U2V2 A0A0N8ERZ1 W5MYM0 A0A182VF85 A0A3P8YN67 A0A195FSV9 F4WFN5 A0A2S2QV01 A0A1S3MAI4 A0A1S3MB54
PDB
3IVF
E-value=6.70946e-18,
Score=215
Ontologies
GO
GO:0005856
GO:0003677
GO:0007016
GO:0051015
GO:0007155
GO:0005925
GO:0001726
GO:0005200
GO:0003779
GO:0043231
GO:0048268
GO:0035615
GO:0035091
GO:0030276
GO:0032051
GO:0043325
GO:0007015
GO:0080025
GO:0030479
GO:0048260
GO:0016021
GO:0014033
GO:0048703
GO:0055013
GO:0007519
GO:0045214
GO:0061386
GO:0007390
GO:0033627
GO:0007475
GO:0086042
GO:0007298
GO:0035160
GO:0045892
GO:0005178
GO:0016203
GO:0007526
GO:0004965
GO:0005891
GO:0043565
GO:0003707
GO:0006281
GO:0006259
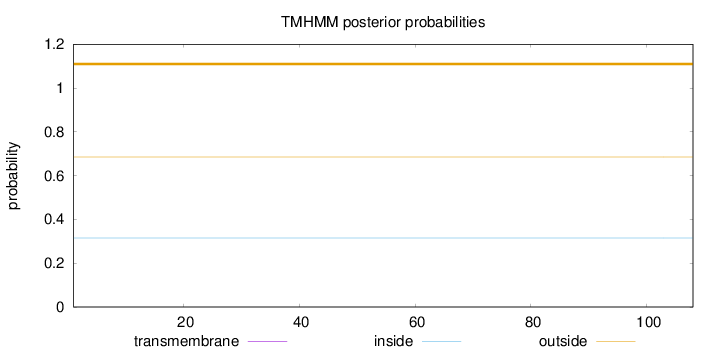
Topology
Length:
108
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.31499
outside
1 - 108
Population Genetic Test Statistics
Pi
137.497782
Theta
141.730401
Tajima's D
0.2531
CLR
7.110675
CSRT
0.44312784360782
Interpretation
Uncertain
