Gene
KWMTBOMO15877 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013369
Annotation
PREDICTED:_vinculin_[Bombyx_mori]
Full name
Vinculin
Location in the cell
Cytoplasmic Reliability : 2.856
Sequence
CDS
ATGCCTGTGTTCCACACGAAAATTATCGAAAGCATTCTAGAGCCAGTCGCTCAGCAGGTGTCTCGACTGGTCATCCTCCATGAAGAAGCAGAAGATGGCAACGCTATGCCGGACTTGGCTCGTCCTGTGCAGGCCGTCTCTCTTGCCGTCAACAACCTCGTCAAGGTTGGCCACGAGACCATAGAGTCATCAGACGATGCGATCCTGCGGCACGACATGCCGGGAGCCTTGCACCGCGTGGAGACGGCAGCCACCCTGCTGCAGCAGGCCTCTGATATGCTCAGAGCTGATCCGTATTCGGGGCCCGCTAGGAAGAAGCTGATCGAAGGGTCCCGCGGTATCCTGCAGGGTACATCGGCGTTGCTCCTCTGTTTCGATGAGTCTGAAGTCAGAAAGATTGTCAAAGAATGCAAGAAGGTTCTAGATTACCTCGGAGTTGCCGAGGTGATAGACACGATGGAGGACTTGGTTCAGTTCCTGAGGGACATCTCGCCGGCGCTCTCCAGGGCGGCCAGAGAGGTGGCGGCCCGCGCTTCGGAGCTGACCCATCCCCCTCACGCCGAGATCCTCACGCAGTGCCTGGAGAGCGTCAAGCAGCTCGCGCCCGTGCTCATCTGCTCCATGAAGATATACATACACATCCTCACCGAGGGCGGCAAGGGCATGGAAGAAGCGGCCGAGAACAGGAACTACCTGGCGCAGAGGATGGCGGATGAGATCCACGAGATCATCAGGGTTCTACAGCTGACGTCGTACGTGGAGGACGGCGGCGAGAAGGACAACATCACCGTGCTTAAAGCTCTCCAAAGTCAGATATACACCAAGATAGGCGCTGCACACGAGTTTTTGAATGACGCGGCGGCGCTGCGCAACAGCTCCGGCGAGCGAGCCCTGCGCTCCGTGCTCACGTCGGCCCAGCGCGCCGCCGAGCACCTGCCGCCCGCGCCCGCCGACGCAGTCGCCGCACGCGTCCGGTGA
Protein
MPVFHTKIIESILEPVAQQVSRLVILHEEAEDGNAMPDLARPVQAVSLAVNNLVKVGHETIESSDDAILRHDMPGALHRVETAATLLQQASDMLRADPYSGPARKKLIEGSRGILQGTSALLLCFDESEVRKIVKECKKVLDYLGVAEVIDTMEDLVQFLRDISPALSRAAREVAARASELTHPPHAEILTQCLESVKQLAPVLICSMKIYIHILTEGGKGMEEAAENRNYLAQRMADEIHEIIRVLQLTSYVEDGGEKDNITVLKALQSQIYTKIGAAHEFLNDAAALRNSSGERALRSVLTSAQRAAEHLPPAPADAVAARVR
Summary
Description
Involved in cell adhesion. May be involved in the attachment of the actin-based microfilaments to the plasma membrane.
Subunit
Exhibits self-association properties.
Similarity
Belongs to the vinculin/alpha-catenin family.
Keywords
Actin-binding
Cell adhesion
Cell junction
Cell membrane
Complete proteome
Cytoplasm
Cytoskeleton
Membrane
Phosphoprotein
Reference proteome
Repeat
Feature
chain Vinculin
Uniprot
A0A1E1W0V8
H9JV06
A0A0N0PCQ9
A0A194PP83
A0A3S2TTN0
A0A212FNK3
+ More
A0A1I8PZF7 A0A0M4FA38 T1PGN4 A0A1Y1KTI0 A0A1A9WVM6 B4NQ05 A0A1A9YMG2 A0A1A9UW50 A0A1B0G8P6 A0A1A9ZKT4 B4L5U0 B4M7C8 A0A3B0JTC8 Q29JL3 T1J8S2 B4Q189 A0A1B6HF25 A0A158NN08 B4JJ77 A0A2J7RNG9 B3P8Y8 A0A151WUQ8 A0A195EWY0 A0A034VPM4 A0A0K8VFK9 X2JAB9 O46037 B3MYS8 A0A195C5N0 A0A026WMK2 A0A1B6G3E0 A0A0A1XKI0 T1HKY3 A0A151JBF5 W8BFM7 A0A3L8DJC6 E2ACE9 A0A1B6ERX3 A0A1L8E3V2 A0A1B0CGY0 Q24584 E2BTN6 A0A069DWV1 A0A2M4A655 A0A2M4BCB1 A0A2M3Z6A1 A0A0A9W3G4 B4I9S2 A0A182FG03 A0A0J7KY03 A0A0A9WQ53 A0A0A9VYZ0 W5JGR3 A0A1W4VS88 A0A1Q3FB41 A0A182NHR3 A0A182PQA9 A0A182X3L7 Q7QJI4 A0A182I3F2 A0A182KRH3 A0A336MF54 A0A131XPW9 A0A1B6CKP3 A0A336MYC4 B0X394 V5GV37 A0A084WNF9 A0A023EWN7 A0A182IJR2 A0A336M9K2 A0A0P4WR17 A0A182JNC5 A0A182WJ44 A0A182Q5G7 Q178I1 A0A182VMB9 A0A131YDG3 V5IK57 A0A131YF27 A0A224Z5F6 A0A0P4WJ01 A0A1E1XF93 A0A0P4W6D5 B7Q5X7 A0A2D3E2W8 U5EZL2 A0A0N7ZCC0 A0A139WG82 A0A226CXN2 A0A1J1HT91 A0A087UMP9 A0A2R5LHL7
A0A1I8PZF7 A0A0M4FA38 T1PGN4 A0A1Y1KTI0 A0A1A9WVM6 B4NQ05 A0A1A9YMG2 A0A1A9UW50 A0A1B0G8P6 A0A1A9ZKT4 B4L5U0 B4M7C8 A0A3B0JTC8 Q29JL3 T1J8S2 B4Q189 A0A1B6HF25 A0A158NN08 B4JJ77 A0A2J7RNG9 B3P8Y8 A0A151WUQ8 A0A195EWY0 A0A034VPM4 A0A0K8VFK9 X2JAB9 O46037 B3MYS8 A0A195C5N0 A0A026WMK2 A0A1B6G3E0 A0A0A1XKI0 T1HKY3 A0A151JBF5 W8BFM7 A0A3L8DJC6 E2ACE9 A0A1B6ERX3 A0A1L8E3V2 A0A1B0CGY0 Q24584 E2BTN6 A0A069DWV1 A0A2M4A655 A0A2M4BCB1 A0A2M3Z6A1 A0A0A9W3G4 B4I9S2 A0A182FG03 A0A0J7KY03 A0A0A9WQ53 A0A0A9VYZ0 W5JGR3 A0A1W4VS88 A0A1Q3FB41 A0A182NHR3 A0A182PQA9 A0A182X3L7 Q7QJI4 A0A182I3F2 A0A182KRH3 A0A336MF54 A0A131XPW9 A0A1B6CKP3 A0A336MYC4 B0X394 V5GV37 A0A084WNF9 A0A023EWN7 A0A182IJR2 A0A336M9K2 A0A0P4WR17 A0A182JNC5 A0A182WJ44 A0A182Q5G7 Q178I1 A0A182VMB9 A0A131YDG3 V5IK57 A0A131YF27 A0A224Z5F6 A0A0P4WJ01 A0A1E1XF93 A0A0P4W6D5 B7Q5X7 A0A2D3E2W8 U5EZL2 A0A0N7ZCC0 A0A139WG82 A0A226CXN2 A0A1J1HT91 A0A087UMP9 A0A2R5LHL7
Pubmed
19121390
26354079
22118469
25315136
28004739
17994087
+ More
15632085 23185243 17550304 21347285 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9280281 10731137 12537569 17372656 24508170 25830018 24495485 30249741 20798317 26334808 25401762 20920257 23761445 12364791 14747013 17210077 20966253 28049606 24438588 24945155 17510324 26830274 25765539 28797301 28503490 18362917 19820115
15632085 23185243 17550304 21347285 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9280281 10731137 12537569 17372656 24508170 25830018 24495485 30249741 20798317 26334808 25401762 20920257 23761445 12364791 14747013 17210077 20966253 28049606 24438588 24945155 17510324 26830274 25765539 28797301 28503490 18362917 19820115
EMBL
GDQN01010424
JAT80630.1
BABH01037485
BABH01037486
BABH01037487
KQ460466
+ More
KPJ14608.1 KQ459597 KPI95246.1 RSAL01000002 RVE54937.1 AGBW02005352 OWR55289.1 CP012528 ALC49537.1 KA647023 AFP61652.1 GEZM01075901 JAV63888.1 CH964291 EDW86230.1 CCAG010000299 CH933811 EDW06549.1 CH940653 EDW62695.1 OUUW01000003 SPP78720.1 CH379063 EAL32288.2 KRT05813.1 JH431963 CM000162 EDX01396.2 GECU01034507 JAS73199.1 ADTU01020808 ADTU01020809 CH916370 EDV99629.1 NEVH01002541 PNF42368.1 CH954183 EDV45593.1 KQ982721 KYQ51650.1 KQ981940 KYN32733.1 GAKP01015203 GAKP01015202 JAC43750.1 GDHF01014667 GDHF01004794 JAI37647.1 JAI47520.1 AE014298 AHN59284.1 AHN59285.1 AL009193 AY128501 CH902632 EDV32772.1 KQ978231 KYM96152.1 KK107152 EZA57138.1 GECZ01012946 JAS56823.1 GBXI01014012 GBXI01012659 GBXI01003214 GBXI01002835 JAD00280.1 JAD01633.1 JAD11078.1 JAD11457.1 ACPB03014658 ACPB03014659 KQ979147 KYN22447.1 GAMC01009048 GAMC01009047 JAB97507.1 QOIP01000007 RLU20520.1 GL438494 EFN68888.1 GECZ01029034 GECZ01018276 GECZ01016277 GECZ01014322 GECZ01013789 GECZ01013540 GECZ01000202 JAS40735.1 JAS51493.1 JAS53492.1 JAS55447.1 JAS55980.1 JAS56229.1 JAS69567.1 GFDF01000780 JAV13304.1 AJWK01011708 X96601 CAA65421.1 GL450467 EFN80951.1 GBGD01000346 JAC88543.1 GGFK01002938 MBW36259.1 GGFJ01001548 MBW50689.1 GGFM01003306 MBW24057.1 GBHO01044200 GBHO01044199 JAF99403.1 JAF99404.1 CH480825 EDW43953.1 LBMM01002163 KMQ95196.1 GBHO01044197 GBHO01036614 JAF99406.1 JAG06990.1 GBHO01044201 GBHO01000457 JAF99402.1 JAG43147.1 ADMH02001234 ETN63552.1 GFDL01010278 JAV24767.1 AAAB01008807 EAA04738.1 APCN01000207 UFQS01000750 UFQT01000750 SSX06610.1 SSX26957.1 GEFH01000970 JAP67611.1 GEDC01023355 GEDC01006906 JAS13943.1 JAS30392.1 UFQT01001504 SSX30858.1 DS232311 EDS39596.1 GALX01000467 JAB67999.1 ATLV01024599 KE525352 KFB51753.1 GAPW01000126 JAC13472.1 SSX06609.1 SSX26956.1 GDRN01069570 JAI64022.1 AXCN02000376 CH477363 EAT42601.1 GEDV01012035 JAP76522.1 GANP01000267 JAB84201.1 GEDV01012036 JAP76521.1 GFPF01013309 MAA24455.1 GDRN01069569 JAI64023.1 GFAC01001251 JAT97937.1 GDRN01069573 JAI64020.1 ABJB010224227 ABJB010232067 ABJB010363238 ABJB010536428 ABJB010621932 ABJB010691707 ABJB010992157 DS863867 EEC14249.1 MF140475 ATU31740.1 GANO01000589 JAB59282.1 GDRN01069568 JAI64024.1 KQ971346 KYB26983.1 LNIX01000061 OXA37247.1 CVRI01000020 CRK91221.1 KK120598 KFM78638.1 GGLE01004888 MBY09014.1
KPJ14608.1 KQ459597 KPI95246.1 RSAL01000002 RVE54937.1 AGBW02005352 OWR55289.1 CP012528 ALC49537.1 KA647023 AFP61652.1 GEZM01075901 JAV63888.1 CH964291 EDW86230.1 CCAG010000299 CH933811 EDW06549.1 CH940653 EDW62695.1 OUUW01000003 SPP78720.1 CH379063 EAL32288.2 KRT05813.1 JH431963 CM000162 EDX01396.2 GECU01034507 JAS73199.1 ADTU01020808 ADTU01020809 CH916370 EDV99629.1 NEVH01002541 PNF42368.1 CH954183 EDV45593.1 KQ982721 KYQ51650.1 KQ981940 KYN32733.1 GAKP01015203 GAKP01015202 JAC43750.1 GDHF01014667 GDHF01004794 JAI37647.1 JAI47520.1 AE014298 AHN59284.1 AHN59285.1 AL009193 AY128501 CH902632 EDV32772.1 KQ978231 KYM96152.1 KK107152 EZA57138.1 GECZ01012946 JAS56823.1 GBXI01014012 GBXI01012659 GBXI01003214 GBXI01002835 JAD00280.1 JAD01633.1 JAD11078.1 JAD11457.1 ACPB03014658 ACPB03014659 KQ979147 KYN22447.1 GAMC01009048 GAMC01009047 JAB97507.1 QOIP01000007 RLU20520.1 GL438494 EFN68888.1 GECZ01029034 GECZ01018276 GECZ01016277 GECZ01014322 GECZ01013789 GECZ01013540 GECZ01000202 JAS40735.1 JAS51493.1 JAS53492.1 JAS55447.1 JAS55980.1 JAS56229.1 JAS69567.1 GFDF01000780 JAV13304.1 AJWK01011708 X96601 CAA65421.1 GL450467 EFN80951.1 GBGD01000346 JAC88543.1 GGFK01002938 MBW36259.1 GGFJ01001548 MBW50689.1 GGFM01003306 MBW24057.1 GBHO01044200 GBHO01044199 JAF99403.1 JAF99404.1 CH480825 EDW43953.1 LBMM01002163 KMQ95196.1 GBHO01044197 GBHO01036614 JAF99406.1 JAG06990.1 GBHO01044201 GBHO01000457 JAF99402.1 JAG43147.1 ADMH02001234 ETN63552.1 GFDL01010278 JAV24767.1 AAAB01008807 EAA04738.1 APCN01000207 UFQS01000750 UFQT01000750 SSX06610.1 SSX26957.1 GEFH01000970 JAP67611.1 GEDC01023355 GEDC01006906 JAS13943.1 JAS30392.1 UFQT01001504 SSX30858.1 DS232311 EDS39596.1 GALX01000467 JAB67999.1 ATLV01024599 KE525352 KFB51753.1 GAPW01000126 JAC13472.1 SSX06609.1 SSX26956.1 GDRN01069570 JAI64022.1 AXCN02000376 CH477363 EAT42601.1 GEDV01012035 JAP76522.1 GANP01000267 JAB84201.1 GEDV01012036 JAP76521.1 GFPF01013309 MAA24455.1 GDRN01069569 JAI64023.1 GFAC01001251 JAT97937.1 GDRN01069573 JAI64020.1 ABJB010224227 ABJB010232067 ABJB010363238 ABJB010536428 ABJB010621932 ABJB010691707 ABJB010992157 DS863867 EEC14249.1 MF140475 ATU31740.1 GANO01000589 JAB59282.1 GDRN01069568 JAI64024.1 KQ971346 KYB26983.1 LNIX01000061 OXA37247.1 CVRI01000020 CRK91221.1 KK120598 KFM78638.1 GGLE01004888 MBY09014.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000095300
+ More
UP000092553 UP000095301 UP000091820 UP000007798 UP000092443 UP000078200 UP000092444 UP000092445 UP000009192 UP000008792 UP000268350 UP000001819 UP000002282 UP000005205 UP000001070 UP000235965 UP000008711 UP000075809 UP000078541 UP000000803 UP000007801 UP000078542 UP000053097 UP000015103 UP000078492 UP000279307 UP000000311 UP000092461 UP000008237 UP000001292 UP000069272 UP000036403 UP000000673 UP000192221 UP000075884 UP000075885 UP000076407 UP000007062 UP000075840 UP000075882 UP000002320 UP000030765 UP000075880 UP000075881 UP000075920 UP000075886 UP000008820 UP000075903 UP000001555 UP000007266 UP000198287 UP000183832 UP000054359
UP000092553 UP000095301 UP000091820 UP000007798 UP000092443 UP000078200 UP000092444 UP000092445 UP000009192 UP000008792 UP000268350 UP000001819 UP000002282 UP000005205 UP000001070 UP000235965 UP000008711 UP000075809 UP000078541 UP000000803 UP000007801 UP000078542 UP000053097 UP000015103 UP000078492 UP000279307 UP000000311 UP000092461 UP000008237 UP000001292 UP000069272 UP000036403 UP000000673 UP000192221 UP000075884 UP000075885 UP000076407 UP000007062 UP000075840 UP000075882 UP000002320 UP000030765 UP000075880 UP000075881 UP000075920 UP000075886 UP000008820 UP000075903 UP000001555 UP000007266 UP000198287 UP000183832 UP000054359
Pfam
PF01044 Vinculin
Interpro
ProteinModelPortal
A0A1E1W0V8
H9JV06
A0A0N0PCQ9
A0A194PP83
A0A3S2TTN0
A0A212FNK3
+ More
A0A1I8PZF7 A0A0M4FA38 T1PGN4 A0A1Y1KTI0 A0A1A9WVM6 B4NQ05 A0A1A9YMG2 A0A1A9UW50 A0A1B0G8P6 A0A1A9ZKT4 B4L5U0 B4M7C8 A0A3B0JTC8 Q29JL3 T1J8S2 B4Q189 A0A1B6HF25 A0A158NN08 B4JJ77 A0A2J7RNG9 B3P8Y8 A0A151WUQ8 A0A195EWY0 A0A034VPM4 A0A0K8VFK9 X2JAB9 O46037 B3MYS8 A0A195C5N0 A0A026WMK2 A0A1B6G3E0 A0A0A1XKI0 T1HKY3 A0A151JBF5 W8BFM7 A0A3L8DJC6 E2ACE9 A0A1B6ERX3 A0A1L8E3V2 A0A1B0CGY0 Q24584 E2BTN6 A0A069DWV1 A0A2M4A655 A0A2M4BCB1 A0A2M3Z6A1 A0A0A9W3G4 B4I9S2 A0A182FG03 A0A0J7KY03 A0A0A9WQ53 A0A0A9VYZ0 W5JGR3 A0A1W4VS88 A0A1Q3FB41 A0A182NHR3 A0A182PQA9 A0A182X3L7 Q7QJI4 A0A182I3F2 A0A182KRH3 A0A336MF54 A0A131XPW9 A0A1B6CKP3 A0A336MYC4 B0X394 V5GV37 A0A084WNF9 A0A023EWN7 A0A182IJR2 A0A336M9K2 A0A0P4WR17 A0A182JNC5 A0A182WJ44 A0A182Q5G7 Q178I1 A0A182VMB9 A0A131YDG3 V5IK57 A0A131YF27 A0A224Z5F6 A0A0P4WJ01 A0A1E1XF93 A0A0P4W6D5 B7Q5X7 A0A2D3E2W8 U5EZL2 A0A0N7ZCC0 A0A139WG82 A0A226CXN2 A0A1J1HT91 A0A087UMP9 A0A2R5LHL7
A0A1I8PZF7 A0A0M4FA38 T1PGN4 A0A1Y1KTI0 A0A1A9WVM6 B4NQ05 A0A1A9YMG2 A0A1A9UW50 A0A1B0G8P6 A0A1A9ZKT4 B4L5U0 B4M7C8 A0A3B0JTC8 Q29JL3 T1J8S2 B4Q189 A0A1B6HF25 A0A158NN08 B4JJ77 A0A2J7RNG9 B3P8Y8 A0A151WUQ8 A0A195EWY0 A0A034VPM4 A0A0K8VFK9 X2JAB9 O46037 B3MYS8 A0A195C5N0 A0A026WMK2 A0A1B6G3E0 A0A0A1XKI0 T1HKY3 A0A151JBF5 W8BFM7 A0A3L8DJC6 E2ACE9 A0A1B6ERX3 A0A1L8E3V2 A0A1B0CGY0 Q24584 E2BTN6 A0A069DWV1 A0A2M4A655 A0A2M4BCB1 A0A2M3Z6A1 A0A0A9W3G4 B4I9S2 A0A182FG03 A0A0J7KY03 A0A0A9WQ53 A0A0A9VYZ0 W5JGR3 A0A1W4VS88 A0A1Q3FB41 A0A182NHR3 A0A182PQA9 A0A182X3L7 Q7QJI4 A0A182I3F2 A0A182KRH3 A0A336MF54 A0A131XPW9 A0A1B6CKP3 A0A336MYC4 B0X394 V5GV37 A0A084WNF9 A0A023EWN7 A0A182IJR2 A0A336M9K2 A0A0P4WR17 A0A182JNC5 A0A182WJ44 A0A182Q5G7 Q178I1 A0A182VMB9 A0A131YDG3 V5IK57 A0A131YF27 A0A224Z5F6 A0A0P4WJ01 A0A1E1XF93 A0A0P4W6D5 B7Q5X7 A0A2D3E2W8 U5EZL2 A0A0N7ZCC0 A0A139WG82 A0A226CXN2 A0A1J1HT91 A0A087UMP9 A0A2R5LHL7
PDB
1ST6
E-value=1.32346e-76,
Score=728
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Cytoplasmic face of adhesion plaques. With evidence from 2 publications.
Cytoskeleton Cytoplasmic face of adhesion plaques. With evidence from 2 publications.
Cell junction Cytoplasmic face of adhesion plaques. With evidence from 2 publications.
Adherens junction Cytoplasmic face of adhesion plaques. With evidence from 2 publications.
Cell membrane Cytoplasmic face of adhesion plaques. With evidence from 2 publications.
Cytoskeleton Cytoplasmic face of adhesion plaques. With evidence from 2 publications.
Cell junction Cytoplasmic face of adhesion plaques. With evidence from 2 publications.
Adherens junction Cytoplasmic face of adhesion plaques. With evidence from 2 publications.
Cell membrane Cytoplasmic face of adhesion plaques. With evidence from 2 publications.
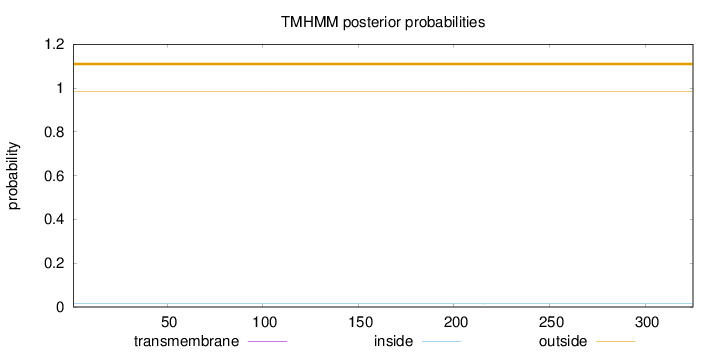
Length:
325
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00188
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01452
outside
1 - 325
Population Genetic Test Statistics
Pi
235.406224
Theta
169.549791
Tajima's D
1.348912
CLR
0.304006
CSRT
0.75691215439228
Interpretation
Uncertain
