Gene
KWMTBOMO15876
Pre Gene Modal
BGIBMGA013368
Annotation
PREDICTED:_gamma-aminobutyric_acid_type_B_receptor_subunit_2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.147 PlasmaMembrane Reliability : 1.883
Sequence
CDS
ATGAAGATGCTTCTCATATTGCTGGTTGTGGTGGCGGTCTGTCGAGGTGTACCTCAGAAGCCGAACCTGGACGCGATACTTAACCGAAGAACCGACGTATACATAGCTGGTTTCTTTCCATTCGGTAAAGGAGTAGAGAATTCCAATACTGGGCGCGGTGTGATGCCGAGTGTGAAGCTAGCCCTGGATCACGTCAACGAACACGACTCGGTGCTGAGGAACTACCGGCTTCACATGTGGTGGAATGACACGGAGTGTAACGCGGCAGTAGGTGTGAAGTCGTTTTTCGACATGATGCACAGCGGTCCTCACAAGCTGATGCTTTTTGGAGCCGCTTGCACGCACGTCACCGATCCCATAGCGAAGGCTTCTAAGCACTGGCATCTTACACAGCTTTCTTACGCCGACACCCACCCGATGTTCACAAGAGAGGCTTTTCCAAATTTCTTCCGCATCGTGCCTTCAGAGAACGCATTCAATTTGCCCCGGATCAGACTACTGCAACACTTCAACTGGACCAGGGTTGGGACTATTTATCAGAACGAACCAAGATATGCTTTGGCTCACAATCGACTACTGGCTGATTTGGACACCCACGGATTCCTGATTGAAGAAACTCAGAGCTTCGCCACTGAAGTAAAAACCGCGTTGGCCAAGCTTAAAGAGAAAGACGTCAGAGTCATTCTTGGAAACTTCAATGAAACCTGGGCCCTGAAGATATTCTGTGAAGCCTACAAACTTGAAATGTTTGGTCGAGCTTACTCCTGGCTCCTGCTGGGGACCTACAGTAACAGCTGGTGGACGCGACGGGCACCTTGTTCCCGCCGGAACTTGACTGCCGCCCTTGACACCGCCATTCTCACGGATCTGCTGCCACTGTCCACCACCCAGGAGACCACCGTTTCTGGCATTACGGCGAAAGACTATCAAGTGGAGTATGACAGGAGACGAGGAATGGAATACTCGAGATTTCACGGCTACACCTACGATGGTATATGGGCGATGGCACTAGCGATACAGACGGTGGCACAACGAGTAAAGCTGAGGAATAAGGAGAAGACAGTGCAGGACTTCCGCTACAGAGACAAGGAGTGGGAGCAGTTGTTCCTGGATGCGCTTAGTAATGTGACATTCGAAGGAGTCACCGGTCCAGTCCGGTTCTATGACAACGAACGCAAAGCCTCTATTCTATTGAAGCAGTTCCAAGGTGATGAAGTAGGGGAAGTGAAGGTGGGGGAGTATTGTGCTGAGAGAGATCATCTGGACCTGACTAGTTGGGAGTCATTCAAATGGCTGGGCAAGAATCCTCCGAAAGACCGAACGCTGCGGCTCATAGAGCACACCCAAGTGAACATCACTATTTACTCTATCTTGGTCTCCTGCTCCGTGCTGGGTATACTCCTAGCCACCGGCTTCCTCGCTATGAACATACATTACAGGAATCAGAGGTACATAAAGATGTCTTCACCGCATTTGAACAATCTCATCATCGTGGGCTGCATGCTCACCTACCTGAGCGTGATCTTCCTCGGCTTGGACTCCAGCCTCAGCAGCATCGCTGCCTTTCCCTACATTTGCACCGCCAGAGCTTGGCTACTAATGGCCGGTTTTAGTCTCGCTTTCGGAGCCATGTTCTCAAAGACTTGGCGGGTCCACTCCATCTTCACTGACGTCAAGCTAAATAAGAAGGTCATTAAAGATTATCAGCTCTTCATGGTCGTGGGAGTGCTTCTCTGCATCGACTTGGCAATAATGACGACTTGGCAGATATCTGATCCTTTTTACAGAGCAACGAAACAGATGGATCCTTACCCTCACCCTTCGAGCGAGGACATCGTGATAGTTCAAGAGAACGAATACTGTCAATCAGAACGGATGCCGATCTTCATTGGAGTCATTTACGCTTACAAGGGCTTGCTGCTGGTATTCGGAGCGTTCCTGGCGTGGGAGACCCGTCATGTCTCTATACCAGCCCTGAACGACTCTCAACACATCGGCTTATCTGTTTACAACGTACTCATCATGTGTATTATGGGCGCAGCTATTACTTTGGTGCTAGCTGACCACAAAGATGCCTTGTTTGTATTAATTGGCATCTTCATAATTTTTTGTACGACGGGAACTCTCTGTTTGGTGTTCGTACCGAAGTTGTTGGAGCTTCGCCGTAACGGTCAAGGCGGGGCGGGGGCGGGACGCATCCGTGCCACACTGCGACCTGCCTCAGGGCACGAGTGCCGCTCGCCCGGACCCGAGCTGGAGAGGCGCCTCAAGGAGCTGAGACAGTACAACAACCGCTACAGGCGGACGTTGCAGGCCAAGGAGAATGAGCTTCAGATGCTACTCAGTAAGCTTGGCAGTGACGTCACTTCGGAATCGTCCACACGCGGAGACTCTCGGTCGCCGGCAACTCAGCGCATAAAGCTGCCCATACCGAAGGAGAATATTAGCCACCCTGAAACAAGCGACATCACGTCTGTGTGCAGTCTGAGCGCCTCAGCTGGCGCGGAAACTGAATACATCAATCTGCAGCCACAACAATCTATCGAGAAACCAAGGCCTTCAGCTCCTCCTGAGCCAATCAAAGACCTAACACCCAAAGAGCCAAAGAAGGAGCAGACGAAAAATGTGTCTTTCAAGAATCATCTGGATTACACCAGTCCTCCGCCTACATCCAAAGCTGTCCCGGAGAAAGATCAGCTCGCGAAATTGCCGTATGGTTGGTCTTCTGAAGTTCGGGAACCATTAGGGCCGGCATTAAAAACATCTAAAACTCCGGAGCCAGCTAAAGCTGAAGCTAAAACCGTACCGGCTCTGAAAAGCAACGACACCCCGCCAGCGATGTCTGTGCCAGCGAAAATCCAGGAGCCGATGTATACCAAGGCAGCCACACCGAAGATCAATAAGACGCCAGAACTTCAAAAGAAAGCTGCTCAAGACTCGGTGTCGACCAAAACAACTCCATCTCGGGAACACAAGCCCGCTAGAGGACATAGACGTATGTCCAGCCTCAGCCAGAGTGCTCTCCTCACTGACCTCATGCATGTCTCCGTGGACCAGGATGATACAGAACCGCCCCCTGGTCTTGAAAGGAAGGTTATAGGTCAAGAAAGAGATCCTCCACCACCTCCAATTGCGAAACCCAAGCAAGAACCGGAAGATATTCTGGACGTAGACCATGATTATTCTTCAATGGAAAGAAGGAAATCTTGCCAAAGAAGAGACTCGGAGAAAAGAAAAAAAGAGAACTTCATAGTAGTCCAGAGCGACCTGTGGGACACTAACACATTTCAGTACACCTGCCAAAAATCTCCTCCAGCCAGTCATCATCACACTCCGATGCAGCGAAGTGTGTCGGAGAAGAGCAGACAGCAGCCTTCACCGCATCACCAAAGGGAAGCGGATAGAACTGTGGAGAGAAGAGCATCAAATGCTTCAATTAATTACGGTCCGGCAAGAGGAGGCACGCCAGAAGGTTATCGAGATGCGGAGACGCTGCCAATGTCTGATCGACATGCCAAGCGTCGAGGATCTGAATTCCCGCAGCCGGTGAGCACTCCTCCACAGCAGTCGCAAACAAAACGAAGACAGTCGATGGCACAGGTGAGATCTTACGCGGAAGAGAGGCCAGATAAAACGGAGATCTCTCGAAGACACGAGGAGCCTAAAAAGAAACAAGAACCTGTCCAGGAATCAGAATGGAAGCCGGAGCCTGATACTATTAGAGTGTCGAAAACCCAGGATTCTCCTGCGAAGAGGCTGAGGCAGGAGGCTGAACCGTTGAAGAGAGCGAAGGCTGTGGAGTCTCCTAGGTTAACCCAGGCCGGCAGTCAGCCTAGGCCAGAACATTACAAGAGCAGCCCTAATGTCGCGTCAAGGCATAAATCTGGCTCACCGAACAAACGCGAAGACAGAAAGTCTGCGTCGTATAGAAGCGATTCAAAGCGTCAAGATTCGGCGGAGCAACGGAAGATGTACACCGCGAGCTCGGAGTCCGAACTGTTCGAATGCGCCATACTCCCTATCTTCCACAAGCTATTGACGGAAAGGCACAAAAGTCAACCGCACGGCTTGAACCTTAGTTACGGGGTCAGCTGTCCTAACATATCCATTAAATGTGACATCGTTGAGTATTTGTAG
Protein
MKMLLILLVVVAVCRGVPQKPNLDAILNRRTDVYIAGFFPFGKGVENSNTGRGVMPSVKLALDHVNEHDSVLRNYRLHMWWNDTECNAAVGVKSFFDMMHSGPHKLMLFGAACTHVTDPIAKASKHWHLTQLSYADTHPMFTREAFPNFFRIVPSENAFNLPRIRLLQHFNWTRVGTIYQNEPRYALAHNRLLADLDTHGFLIEETQSFATEVKTALAKLKEKDVRVILGNFNETWALKIFCEAYKLEMFGRAYSWLLLGTYSNSWWTRRAPCSRRNLTAALDTAILTDLLPLSTTQETTVSGITAKDYQVEYDRRRGMEYSRFHGYTYDGIWAMALAIQTVAQRVKLRNKEKTVQDFRYRDKEWEQLFLDALSNVTFEGVTGPVRFYDNERKASILLKQFQGDEVGEVKVGEYCAERDHLDLTSWESFKWLGKNPPKDRTLRLIEHTQVNITIYSILVSCSVLGILLATGFLAMNIHYRNQRYIKMSSPHLNNLIIVGCMLTYLSVIFLGLDSSLSSIAAFPYICTARAWLLMAGFSLAFGAMFSKTWRVHSIFTDVKLNKKVIKDYQLFMVVGVLLCIDLAIMTTWQISDPFYRATKQMDPYPHPSSEDIVIVQENEYCQSERMPIFIGVIYAYKGLLLVFGAFLAWETRHVSIPALNDSQHIGLSVYNVLIMCIMGAAITLVLADHKDALFVLIGIFIIFCTTGTLCLVFVPKLLELRRNGQGGAGAGRIRATLRPASGHECRSPGPELERRLKELRQYNNRYRRTLQAKENELQMLLSKLGSDVTSESSTRGDSRSPATQRIKLPIPKENISHPETSDITSVCSLSASAGAETEYINLQPQQSIEKPRPSAPPEPIKDLTPKEPKKEQTKNVSFKNHLDYTSPPPTSKAVPEKDQLAKLPYGWSSEVREPLGPALKTSKTPEPAKAEAKTVPALKSNDTPPAMSVPAKIQEPMYTKAATPKINKTPELQKKAAQDSVSTKTTPSREHKPARGHRRMSSLSQSALLTDLMHVSVDQDDTEPPPGLERKVIGQERDPPPPPIAKPKQEPEDILDVDHDYSSMERRKSCQRRDSEKRKKENFIVVQSDLWDTNTFQYTCQKSPPASHHHTPMQRSVSEKSRQQPSPHHQREADRTVERRASNASINYGPARGGTPEGYRDAETLPMSDRHAKRRGSEFPQPVSTPPQQSQTKRRQSMAQVRSYAEERPDKTEISRRHEEPKKKQEPVQESEWKPEPDTIRVSKTQDSPAKRLRQEAEPLKRAKAVESPRLTQAGSQPRPEHYKSSPNVASRHKSGSPNKREDRKSASYRSDSKRQDSAEQRKMYTASSESELFECAILPIFHKLLTERHKSQPHGLNLSYGVSCPNISIKCDIVEYL
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF53822
SSF53822
ProteinModelPortal
PDB
4MS4
E-value=1.12001e-80,
Score=769
Ontologies
GO
PANTHER
Topology
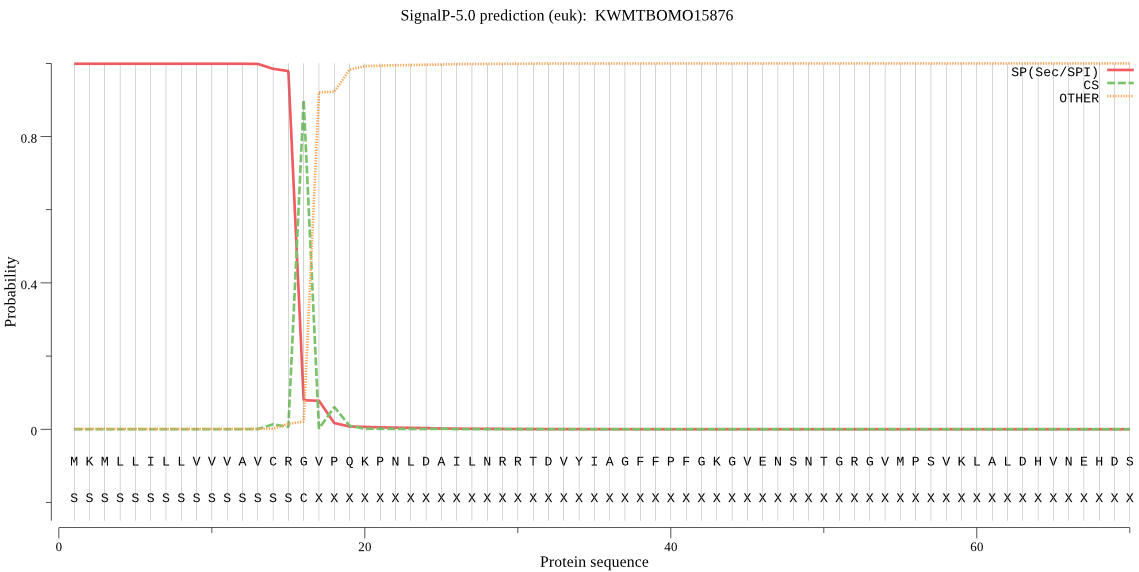
SignalP
Position: 1 - 16,
Likelihood: 0.998718
Length:
1378
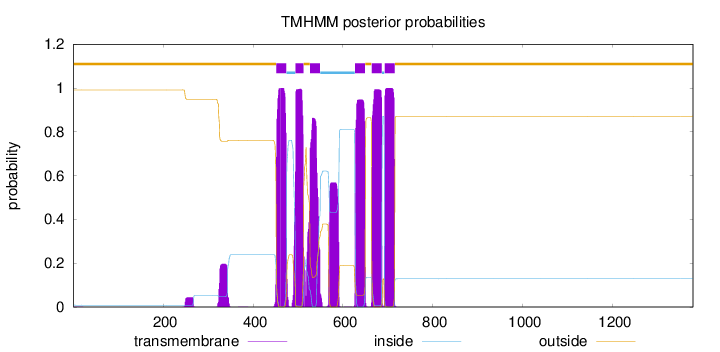
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
146.33192
Exp number, first 60 AAs:
0.01665
Total prob of N-in:
0.00911
outside
1 - 451
TMhelix
452 - 474
inside
475 - 494
TMhelix
495 - 512
outside
513 - 526
TMhelix
527 - 549
inside
550 - 626
TMhelix
627 - 649
outside
650 - 663
TMhelix
664 - 686
inside
687 - 692
TMhelix
693 - 715
outside
716 - 1378
Population Genetic Test Statistics
Pi
223.258354
Theta
159.662879
Tajima's D
0.602165
CLR
0.661377
CSRT
0.548122593870306
Interpretation
Uncertain
