Gene
KWMTBOMO15870 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013348
Annotation
PREDICTED:_mitochondrial_import_receptor_subunit_TOM40_homolog_[Papilio_polytes]
Full name
Serine/threonine-protein kinase TOR
Location in the cell
Cytoplasmic Reliability : 1.324 Mitochondrial Reliability : 1.151
Sequence
CDS
ATGGGTGCTATATCAAGCAGGTACCGTGTCTTCAACTCTGGGCTGACTAGTACCGACTTACAAAATATAGAAAATGATAACAAACGAGAAAATCCAGGGACTTTGGATGAACTTCATAAAAAAACCAAAGAGGTGATGCCTGTGAACTTTGAAGGTGGCAAGTTAATGGTCAATAGAGGGCTGTCAAATCATTTTCAGATGTCTCATACATTAACAATGAGCTCACAACAAAATGGATACAAATTGGGTGGAACATACATTGGCACAAAACAGATATCTCCGACAGAGGCATTCCCGGTGGTGCTTGGTGATGTGGACCCTGCCGGTAATGTGAACTGCAGCCTTATACATCAACTGACACCAGAGATCAGGGTCAAAGGTGCTGCTCAAATTCAAGAATGCAAACTGACAGCTACACAAGGCACTGTGGAATACAAAGGTTCAGATTACAGCCTTGCCTTGGTGGCGGGGAAACCAGACTTTAACGACAAGTCTGCAATGTTTGTTGGACATTATTTGCAGTCGGTGACAAAGCGTTTAGCTTTAGGGGCTGAACTGGTGTATCAATCCGGTGCCAGGATCATGGGCGGAGAGATTTGCATAGTGTCTGGTGCTGCTAGATACACTATGGATGATTCAGAAGTGTCGGCCACACTGAGCGCTGCCAACCTGCACTTGTGCTACTTCAAGCAAGCCAGCGAACAACTCCAGGTTGTAGCTGAAATGGACACATCATTCCGTGGCATGGAGTCAGTGGGCAGCATCGGATATCAAGTCACTATTCCAAAGGCAGATCTAGTTTTTAGAGGCATGGTGGATTCCAACTGGAACATCGGAGCAGTCTTGGAGAAGAAACTCCAGCCCCTGCCCTTCACGTTCGCGCTGTCGGGCATGGCCAACCAGGCCAAGCAGCAGTTCAAGTTTGGCTGCGGACTTATCATCGGTTAA
Protein
MGAISSRYRVFNSGLTSTDLQNIENDNKRENPGTLDELHKKTKEVMPVNFEGGKLMVNRGLSNHFQMSHTLTMSSQQNGYKLGGTYIGTKQISPTEAFPVVLGDVDPAGNVNCSLIHQLTPEIRVKGAAQIQECKLTATQGTVEYKGSDYSLALVAGKPDFNDKSAMFVGHYLQSVTKRLALGAELVYQSGARIMGGEICIVSGAARYTMDDSEVSATLSAANLHLCYFKQASEQLQVVAEMDTSFRGMESVGSIGYQVTIPKADLVFRGMVDSNWNIGAVLEKKLQPLPFTFALSGMANQAKQQFKFGCGLIIG
Summary
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Similarity
Belongs to the PI3/PI4-kinase family.
Uniprot
H9JUY5
A0A3S2M0K9
A0A2H1WBQ1
A0A2A4JF87
A0A194PQJ6
A0A212F4R4
+ More
A0A1E1WTS0 A0A0N1PJG0 A0A182QS84 A0A182LYZ7 A0A182L298 Q7Q3H1 A0A182TNI8 A0A084VP73 A0A182UMC9 A0A182HVD6 A0A182NE95 A0A182JTH9 A0A182J1Y3 A0A182YGZ4 A0A182W9V5 A0A2M4BTH6 A0A182PMB0 A0A2M3ZGV6 A0A2M3Z3B3 A0A2M4AHZ8 A0A336LRW8 W5J8U5 A0A1A9WH28 A0A1A9VBP9 T1D4E1 A0A1A9Z632 A0A1B0G8B9 A0A182H6N8 B0W677 A0A1A9XXR4 A0A1B0APG7 A0A023EPM8 Q173U9 A0A034WE23 A0A1Q3F4E0 A0A0K8TLU0 A0A0K8VCU3 A0A2J7R264 A0A0L0BTN3 A0A1I8PBG7 T1PD68 A0A0M3QZ45 A0A2R5LBV8 A0A1E1XQK4 A0A0P5FC59 A0A0A1XEU8 E9GS42 A0A131Z4J4 A0A224ZAK4 L7M801 A0A023GLL5 G3MM40 A0A0P6C3N8 A0A1E1XFF4 W8C5C2 V5HQS4 A0A182T030 B4L6B8 A0A1Z5KVY8 A0A232FGG1 A0A293MYD5 K7IPD3 B4MAF0 A0A0P5BQB7 A0A151I2R9 A0A158NVQ0 B4NCX2 B4JK31 V9IIB9 A0A088AHF2 A0A151IDN2 A0A131XDL3 A0A0N0U2Z7 A0A151WVS7 E2A3T5 F4X7U5 E9J902 A0A0J7L0V2 A0A151JAL4 A0A3B0JYA4 D6WGH1 A0A165A1G4 A0A195FXF0 A0A026WQS2 A0A1Y1LXH6 B4GVU8 A0A310SAW1 Q29G95 A0A182WTE2 A0A067R567 B3NTS7 B4Q0S9 A0A1W4VK16
A0A1E1WTS0 A0A0N1PJG0 A0A182QS84 A0A182LYZ7 A0A182L298 Q7Q3H1 A0A182TNI8 A0A084VP73 A0A182UMC9 A0A182HVD6 A0A182NE95 A0A182JTH9 A0A182J1Y3 A0A182YGZ4 A0A182W9V5 A0A2M4BTH6 A0A182PMB0 A0A2M3ZGV6 A0A2M3Z3B3 A0A2M4AHZ8 A0A336LRW8 W5J8U5 A0A1A9WH28 A0A1A9VBP9 T1D4E1 A0A1A9Z632 A0A1B0G8B9 A0A182H6N8 B0W677 A0A1A9XXR4 A0A1B0APG7 A0A023EPM8 Q173U9 A0A034WE23 A0A1Q3F4E0 A0A0K8TLU0 A0A0K8VCU3 A0A2J7R264 A0A0L0BTN3 A0A1I8PBG7 T1PD68 A0A0M3QZ45 A0A2R5LBV8 A0A1E1XQK4 A0A0P5FC59 A0A0A1XEU8 E9GS42 A0A131Z4J4 A0A224ZAK4 L7M801 A0A023GLL5 G3MM40 A0A0P6C3N8 A0A1E1XFF4 W8C5C2 V5HQS4 A0A182T030 B4L6B8 A0A1Z5KVY8 A0A232FGG1 A0A293MYD5 K7IPD3 B4MAF0 A0A0P5BQB7 A0A151I2R9 A0A158NVQ0 B4NCX2 B4JK31 V9IIB9 A0A088AHF2 A0A151IDN2 A0A131XDL3 A0A0N0U2Z7 A0A151WVS7 E2A3T5 F4X7U5 E9J902 A0A0J7L0V2 A0A151JAL4 A0A3B0JYA4 D6WGH1 A0A165A1G4 A0A195FXF0 A0A026WQS2 A0A1Y1LXH6 B4GVU8 A0A310SAW1 Q29G95 A0A182WTE2 A0A067R567 B3NTS7 B4Q0S9 A0A1W4VK16
EC Number
2.7.11.1
Pubmed
19121390
26354079
22118469
20966253
12364791
14747013
+ More
17210077 24438588 25244985 20920257 23761445 24330624 26483478 24945155 17510324 25348373 26369729 26108605 25315136 29209593 25830018 21292972 26830274 28797301 25576852 22216098 28503490 24495485 25765539 17994087 28528879 28648823 20075255 21347285 28049606 20798317 21719571 21282665 18362917 19820115 24508170 30249741 28004739 15632085 24845553 17550304
17210077 24438588 25244985 20920257 23761445 24330624 26483478 24945155 17510324 25348373 26369729 26108605 25315136 29209593 25830018 21292972 26830274 28797301 25576852 22216098 28503490 24495485 25765539 17994087 28528879 28648823 20075255 21347285 28049606 20798317 21719571 21282665 18362917 19820115 24508170 30249741 28004739 15632085 24845553 17550304
EMBL
BABH01037461
BABH01037462
RSAL01000085
RVE48332.1
ODYU01007597
SOQ50515.1
+ More
NWSH01001597 PCG70755.1 KQ459597 KPI95238.1 AGBW02010326 OWR48713.1 GDQN01000621 JAT90433.1 KQ460466 KPJ14617.1 AXCN02000864 AXCM01000972 AAAB01008964 EAA12891.2 ATLV01014983 KE524999 KFB39767.1 APCN01002505 GGFJ01007173 MBW56314.1 GGFM01006917 MBW27668.1 GGFM01002253 MBW23004.1 GGFK01007080 MBW40401.1 UFQT01000140 SSX20734.1 ADMH02002093 ETN59300.1 GALA01000993 JAA93859.1 CCAG010000229 JXUM01114672 JXUM01114673 JXUM01114674 JXUM01114675 JXUM01114676 JXUM01114677 JXUM01114678 JXUM01114679 KQ565807 KXJ70650.1 DS231847 EDS36424.1 JXJN01001399 GAPW01002638 JAC10960.1 CH477416 EAT41347.1 GAKP01006013 JAC52939.1 GFDL01012656 JAV22389.1 GDAI01002698 JAI14905.1 GDHF01015575 JAI36739.1 NEVH01008200 PNF34925.1 JRES01001480 KNC22589.1 KA646060 AFP60689.1 CP012528 ALC48729.1 GGLE01002877 MBY07003.1 GFAA01001847 JAU01588.1 GDIP01245662 GDIQ01257220 JAI77739.1 JAJ94504.1 GBXI01004805 JAD09487.1 GL732561 EFX77643.1 GEDV01002234 JAP86323.1 GFPF01012758 MAA23904.1 GACK01004613 JAA60421.1 GBBM01001708 JAC33710.1 JO842941 AEO34558.1 GDIP01021531 JAM82184.1 GFAC01001198 JAT97990.1 GAMC01004959 JAC01597.1 GANP01008145 JAB76323.1 CH933812 EDW05914.1 GFJQ02007750 JAV99219.1 NNAY01000229 OXU29856.1 GFWV01019622 MAA44350.1 CH940655 EDW66209.1 GDIP01186152 JAJ37250.1 KQ976554 KYM80822.1 ADTU01027371 CH964239 EDW82681.1 CH916370 EDV99933.1 JR049168 AEY60883.1 KQ977979 KYM98359.1 GEFH01004825 JAP63756.1 KQ435969 KOX67844.1 KQ982709 KYQ51775.1 GL436478 EFN71901.1 GL888900 EGI57431.1 GL769168 EFZ10669.1 LBMM01001492 KMQ96218.1 KQ979254 KYN22105.1 OUUW01000003 SPP78346.1 KQ971321 EFA00558.1 LRGB01000660 KZS17075.1 KQ981193 KYN45113.1 KK107128 QOIP01000006 EZA58298.1 RLU21625.1 GEZM01046785 GEZM01046784 JAV77010.1 CH479193 EDW26793.1 KQ775960 OAD52241.1 CH379064 EAL32215.1 KK852959 KDR13229.1 CH954180 EDV47490.1 CM000162 EDX02350.1 KRK06720.1
NWSH01001597 PCG70755.1 KQ459597 KPI95238.1 AGBW02010326 OWR48713.1 GDQN01000621 JAT90433.1 KQ460466 KPJ14617.1 AXCN02000864 AXCM01000972 AAAB01008964 EAA12891.2 ATLV01014983 KE524999 KFB39767.1 APCN01002505 GGFJ01007173 MBW56314.1 GGFM01006917 MBW27668.1 GGFM01002253 MBW23004.1 GGFK01007080 MBW40401.1 UFQT01000140 SSX20734.1 ADMH02002093 ETN59300.1 GALA01000993 JAA93859.1 CCAG010000229 JXUM01114672 JXUM01114673 JXUM01114674 JXUM01114675 JXUM01114676 JXUM01114677 JXUM01114678 JXUM01114679 KQ565807 KXJ70650.1 DS231847 EDS36424.1 JXJN01001399 GAPW01002638 JAC10960.1 CH477416 EAT41347.1 GAKP01006013 JAC52939.1 GFDL01012656 JAV22389.1 GDAI01002698 JAI14905.1 GDHF01015575 JAI36739.1 NEVH01008200 PNF34925.1 JRES01001480 KNC22589.1 KA646060 AFP60689.1 CP012528 ALC48729.1 GGLE01002877 MBY07003.1 GFAA01001847 JAU01588.1 GDIP01245662 GDIQ01257220 JAI77739.1 JAJ94504.1 GBXI01004805 JAD09487.1 GL732561 EFX77643.1 GEDV01002234 JAP86323.1 GFPF01012758 MAA23904.1 GACK01004613 JAA60421.1 GBBM01001708 JAC33710.1 JO842941 AEO34558.1 GDIP01021531 JAM82184.1 GFAC01001198 JAT97990.1 GAMC01004959 JAC01597.1 GANP01008145 JAB76323.1 CH933812 EDW05914.1 GFJQ02007750 JAV99219.1 NNAY01000229 OXU29856.1 GFWV01019622 MAA44350.1 CH940655 EDW66209.1 GDIP01186152 JAJ37250.1 KQ976554 KYM80822.1 ADTU01027371 CH964239 EDW82681.1 CH916370 EDV99933.1 JR049168 AEY60883.1 KQ977979 KYM98359.1 GEFH01004825 JAP63756.1 KQ435969 KOX67844.1 KQ982709 KYQ51775.1 GL436478 EFN71901.1 GL888900 EGI57431.1 GL769168 EFZ10669.1 LBMM01001492 KMQ96218.1 KQ979254 KYN22105.1 OUUW01000003 SPP78346.1 KQ971321 EFA00558.1 LRGB01000660 KZS17075.1 KQ981193 KYN45113.1 KK107128 QOIP01000006 EZA58298.1 RLU21625.1 GEZM01046785 GEZM01046784 JAV77010.1 CH479193 EDW26793.1 KQ775960 OAD52241.1 CH379064 EAL32215.1 KK852959 KDR13229.1 CH954180 EDV47490.1 CM000162 EDX02350.1 KRK06720.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000053240
+ More
UP000075886 UP000075883 UP000075882 UP000007062 UP000075902 UP000030765 UP000075903 UP000075840 UP000075884 UP000075881 UP000075880 UP000076408 UP000075920 UP000075885 UP000000673 UP000091820 UP000078200 UP000092445 UP000092444 UP000069940 UP000249989 UP000002320 UP000092443 UP000092460 UP000008820 UP000235965 UP000037069 UP000095300 UP000095301 UP000092553 UP000000305 UP000075901 UP000009192 UP000215335 UP000002358 UP000008792 UP000078540 UP000005205 UP000007798 UP000001070 UP000005203 UP000078542 UP000053105 UP000075809 UP000000311 UP000007755 UP000036403 UP000078492 UP000268350 UP000007266 UP000076858 UP000078541 UP000053097 UP000279307 UP000008744 UP000001819 UP000076407 UP000027135 UP000008711 UP000002282 UP000192221
UP000075886 UP000075883 UP000075882 UP000007062 UP000075902 UP000030765 UP000075903 UP000075840 UP000075884 UP000075881 UP000075880 UP000076408 UP000075920 UP000075885 UP000000673 UP000091820 UP000078200 UP000092445 UP000092444 UP000069940 UP000249989 UP000002320 UP000092443 UP000092460 UP000008820 UP000235965 UP000037069 UP000095300 UP000095301 UP000092553 UP000000305 UP000075901 UP000009192 UP000215335 UP000002358 UP000008792 UP000078540 UP000005205 UP000007798 UP000001070 UP000005203 UP000078542 UP000053105 UP000075809 UP000000311 UP000007755 UP000036403 UP000078492 UP000268350 UP000007266 UP000076858 UP000078541 UP000053097 UP000279307 UP000008744 UP000001819 UP000076407 UP000027135 UP000008711 UP000002282 UP000192221
PRIDE
Pfam
Interpro
IPR037930
Tom40
+ More
IPR027246 Porin_Euk/Tom40
IPR023614 Porin_dom_sf
IPR024585 DUF3385_TOR
IPR036738 FRB_sf
IPR036940 PI3/4_kinase_cat_sf
IPR026683 TOR
IPR009076 FRB_dom
IPR003152 FATC_dom
IPR018936 PI3/4_kinase_CS
IPR000403 PI3/4_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR011990 TPR-like_helical_dom_sf
IPR016024 ARM-type_fold
IPR003151 PIK-rel_kinase_FAT
IPR011989 ARM-like
IPR014009 PIK_FAT
IPR027246 Porin_Euk/Tom40
IPR023614 Porin_dom_sf
IPR024585 DUF3385_TOR
IPR036738 FRB_sf
IPR036940 PI3/4_kinase_cat_sf
IPR026683 TOR
IPR009076 FRB_dom
IPR003152 FATC_dom
IPR018936 PI3/4_kinase_CS
IPR000403 PI3/4_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR011990 TPR-like_helical_dom_sf
IPR016024 ARM-type_fold
IPR003151 PIK-rel_kinase_FAT
IPR011989 ARM-like
IPR014009 PIK_FAT
ProteinModelPortal
H9JUY5
A0A3S2M0K9
A0A2H1WBQ1
A0A2A4JF87
A0A194PQJ6
A0A212F4R4
+ More
A0A1E1WTS0 A0A0N1PJG0 A0A182QS84 A0A182LYZ7 A0A182L298 Q7Q3H1 A0A182TNI8 A0A084VP73 A0A182UMC9 A0A182HVD6 A0A182NE95 A0A182JTH9 A0A182J1Y3 A0A182YGZ4 A0A182W9V5 A0A2M4BTH6 A0A182PMB0 A0A2M3ZGV6 A0A2M3Z3B3 A0A2M4AHZ8 A0A336LRW8 W5J8U5 A0A1A9WH28 A0A1A9VBP9 T1D4E1 A0A1A9Z632 A0A1B0G8B9 A0A182H6N8 B0W677 A0A1A9XXR4 A0A1B0APG7 A0A023EPM8 Q173U9 A0A034WE23 A0A1Q3F4E0 A0A0K8TLU0 A0A0K8VCU3 A0A2J7R264 A0A0L0BTN3 A0A1I8PBG7 T1PD68 A0A0M3QZ45 A0A2R5LBV8 A0A1E1XQK4 A0A0P5FC59 A0A0A1XEU8 E9GS42 A0A131Z4J4 A0A224ZAK4 L7M801 A0A023GLL5 G3MM40 A0A0P6C3N8 A0A1E1XFF4 W8C5C2 V5HQS4 A0A182T030 B4L6B8 A0A1Z5KVY8 A0A232FGG1 A0A293MYD5 K7IPD3 B4MAF0 A0A0P5BQB7 A0A151I2R9 A0A158NVQ0 B4NCX2 B4JK31 V9IIB9 A0A088AHF2 A0A151IDN2 A0A131XDL3 A0A0N0U2Z7 A0A151WVS7 E2A3T5 F4X7U5 E9J902 A0A0J7L0V2 A0A151JAL4 A0A3B0JYA4 D6WGH1 A0A165A1G4 A0A195FXF0 A0A026WQS2 A0A1Y1LXH6 B4GVU8 A0A310SAW1 Q29G95 A0A182WTE2 A0A067R567 B3NTS7 B4Q0S9 A0A1W4VK16
A0A1E1WTS0 A0A0N1PJG0 A0A182QS84 A0A182LYZ7 A0A182L298 Q7Q3H1 A0A182TNI8 A0A084VP73 A0A182UMC9 A0A182HVD6 A0A182NE95 A0A182JTH9 A0A182J1Y3 A0A182YGZ4 A0A182W9V5 A0A2M4BTH6 A0A182PMB0 A0A2M3ZGV6 A0A2M3Z3B3 A0A2M4AHZ8 A0A336LRW8 W5J8U5 A0A1A9WH28 A0A1A9VBP9 T1D4E1 A0A1A9Z632 A0A1B0G8B9 A0A182H6N8 B0W677 A0A1A9XXR4 A0A1B0APG7 A0A023EPM8 Q173U9 A0A034WE23 A0A1Q3F4E0 A0A0K8TLU0 A0A0K8VCU3 A0A2J7R264 A0A0L0BTN3 A0A1I8PBG7 T1PD68 A0A0M3QZ45 A0A2R5LBV8 A0A1E1XQK4 A0A0P5FC59 A0A0A1XEU8 E9GS42 A0A131Z4J4 A0A224ZAK4 L7M801 A0A023GLL5 G3MM40 A0A0P6C3N8 A0A1E1XFF4 W8C5C2 V5HQS4 A0A182T030 B4L6B8 A0A1Z5KVY8 A0A232FGG1 A0A293MYD5 K7IPD3 B4MAF0 A0A0P5BQB7 A0A151I2R9 A0A158NVQ0 B4NCX2 B4JK31 V9IIB9 A0A088AHF2 A0A151IDN2 A0A131XDL3 A0A0N0U2Z7 A0A151WVS7 E2A3T5 F4X7U5 E9J902 A0A0J7L0V2 A0A151JAL4 A0A3B0JYA4 D6WGH1 A0A165A1G4 A0A195FXF0 A0A026WQS2 A0A1Y1LXH6 B4GVU8 A0A310SAW1 Q29G95 A0A182WTE2 A0A067R567 B3NTS7 B4Q0S9 A0A1W4VK16
PDB
5O8O
E-value=3.24341e-16,
Score=207
Ontologies
GO
PANTHER
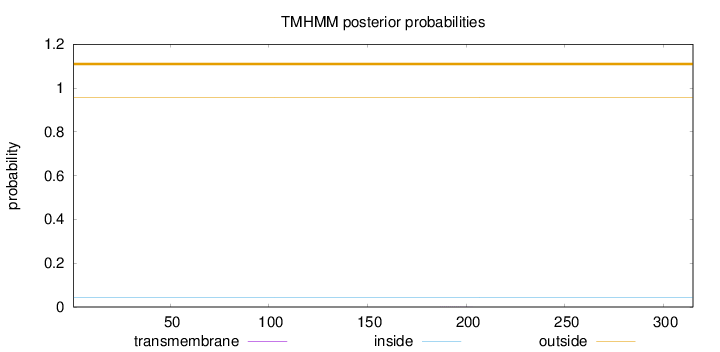
Topology
Length:
315
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00495999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04482
outside
1 - 315
Population Genetic Test Statistics
Pi
213.770113
Theta
170.324259
Tajima's D
1.041207
CLR
0.011326
CSRT
0.67651617419129
Interpretation
Uncertain
