Gene
KWMTBOMO15864
Pre Gene Modal
BGIBMGA013352
Annotation
PREDICTED:_nuclear_factor_1_B-type_[Bombyx_mori]
Full name
Nuclear factor 1
Transcription factor
Location in the cell
Nuclear Reliability : 4.107
Sequence
CDS
ATGTTATACGGCTATATACGAAAGGACGAGTTCCATCCGTTCATAGAGGCCCTAATGCCTTTCGTTAAATCGTTTTCGTACACGTGGTTCAATTTACAAGCGGCGAAACGGAAATATTACAAAAAACACGAGAAACGGATGAGTCTTGAGGAGGAGCGGCATACTAAATATGAATTACAGAATGAAAAAGCAGAAGTCAAACAAAAATGGGCATCCCGGTTACTGGGAAAGCTTCGGAAGGACATCACCCAAGACTGCAGAGAGGACTTCGTGCTCAGCATCACGGGCAAGAGGCCGGCGGTCTGTGTCCTCTCCAACCCGGACCAGAAGGGCAAGATGCGGAGGATAGACTGCTTGAGGCAGGCCGATAAGGTGTGGAGACTAGATCTGGTCATGGTGATACTCTTCAAAGCCATACCCTTGGAGAGCACGGACGGAGAGCGACTAGAGAAGCACCCTGAGTGTACGCAGCCAGGTCTCTGCGTGAACCCGTACCACATCAACGTCTCTGTCCGCGAGCTGGACCTCTACCTCGCAAACTTCATCAACAGCTATGATATCCTAAGTGGGTCCATCTCTCCTCATCCACCTAGAGACAAAGAGAACGAACACGAGGCTAAGAACAAAGGTTATAGTCACAATCCATACAATGGTGTTATTTGCAACGACATAATACTAGCGACGGGTGTCTTCTCTTCTAAGGAACTATGGAAGTTGTCTAAAGCGTCAATCCTGCAAGAGACCGGTTCTGAATCACCTGGTGGTTCCGGTGGTGGTATCAAGTTGGAATCGGGATATTACTGCGGGTACAATAGTCCAGCACCACCCACCTTCGATCCTCCGGCGGAGAGGTCCACTCCGTCGACCATGCTTGTCGGGCAGTGCTTTAGTATCCCCCACAGCTCAGCGGGTCTGAGTTCGGCCCAGTCGCCGCTGGTGCCGTCCAACACGATATTCTACCAGCCGCCGCCCGCGGACACGCACCCCAATACATCAGAAGCCTTAACGAGTCATCATTCGAGCGCGAAGTACGAGACGCCGCAGGACTCTCTCGGGGACTTCGTGACCTTCGTCTGCCAGGAGCCAACAGATGTTGCTCAATTACAGGTTCATAGCTTGTCGAGGAGTCCTAAGCCGGCGTACTTCAGCAGCTCGATGCTACCCCCACCCCCCCTACCCCCGATGGCGAGACCAGTAGCTATCATTAGATCGACAGGTATTTTTTGA
Protein
MLYGYIRKDEFHPFIEALMPFVKSFSYTWFNLQAAKRKYYKKHEKRMSLEEERHTKYELQNEKAEVKQKWASRLLGKLRKDITQDCREDFVLSITGKRPAVCVLSNPDQKGKMRRIDCLRQADKVWRLDLVMVILFKAIPLESTDGERLEKHPECTQPGLCVNPYHINVSVRELDLYLANFINSYDILSGSISPHPPRDKENEHEAKNKGYSHNPYNGVICNDIILATGVFSSKELWKLSKASILQETGSESPGGSGGGIKLESGYYCGYNSPAPPTFDPPAERSTPSTMLVGQCFSIPHSSAGLSSAQSPLVPSNTIFYQPPPADTHPNTSEALTSHHSSAKYETPQDSLGDFVTFVCQEPTDVAQLQVHSLSRSPKPAYFSSSMLPPPPLPPMARPVAIIRSTGIF
Summary
Description
Recognizes and binds the palindromic sequence 5'-TTGGCNNNNNGCCAA-3' present in viral and cellular promoters and in the origin of replication of adenovirus type 2. These proteins are individually capable of activating transcription and replication.
Subunit
Binds DNA as a homodimer.
Similarity
Belongs to the CTF/NF-I family.
Uniprot
H9JUY9
A0A2A4J1F4
A0A2A4J301
A0A2A4J340
A0A2A4J2T0
A0A2A4J351
+ More
A0A2H1W5W7 A0A0N0PCV7 A0A194PP70 A0A0L7LPP8 A0A1W4WJL8 A0A1W4WKK1 A0A1W4WJL3 A0A1Y1N493 A0A1Y1N7K5 D6W8F2 N6T9B3 A0A1B6JCB9 A0A1B6GS96 A0A1B6K198 A0A1B6H2C3 A0A1B6CGE6 X1WJN8 A0A1B6EH04 A0A1J1J9P0 A0A1B6LVR4 A0A1B6GL89 A0A0K8SRB8 A0A1B6G839 A0A1B6JW54 A0A1B6M9R2 A0A195C3F0 A0A1B6F7Q4 A0A0A9WEI4 A0A1B6J430 A0A1B6BYV1 A0A151WGI1 A0A146LGC7 A0A067RDG1 A0A154P719 A0A2S2PHH2 A0A026WS15 A0A2S2NT65 A0A1B6DXH0 E2AW03 A0A1W4W9I4 A0A1W4WKU2 A0A1B6L878 A0A0T6B0N5 A0A1B6KH13 W8BNL0 A0A336KYY0 K7J354 A0A0P8XII8 B3N0M1 E2C4L7
A0A2H1W5W7 A0A0N0PCV7 A0A194PP70 A0A0L7LPP8 A0A1W4WJL8 A0A1W4WKK1 A0A1W4WJL3 A0A1Y1N493 A0A1Y1N7K5 D6W8F2 N6T9B3 A0A1B6JCB9 A0A1B6GS96 A0A1B6K198 A0A1B6H2C3 A0A1B6CGE6 X1WJN8 A0A1B6EH04 A0A1J1J9P0 A0A1B6LVR4 A0A1B6GL89 A0A0K8SRB8 A0A1B6G839 A0A1B6JW54 A0A1B6M9R2 A0A195C3F0 A0A1B6F7Q4 A0A0A9WEI4 A0A1B6J430 A0A1B6BYV1 A0A151WGI1 A0A146LGC7 A0A067RDG1 A0A154P719 A0A2S2PHH2 A0A026WS15 A0A2S2NT65 A0A1B6DXH0 E2AW03 A0A1W4W9I4 A0A1W4WKU2 A0A1B6L878 A0A0T6B0N5 A0A1B6KH13 W8BNL0 A0A336KYY0 K7J354 A0A0P8XII8 B3N0M1 E2C4L7
Pubmed
EMBL
BABH01037450
BABH01037451
NWSH01003786
PCG65821.1
PCG65820.1
PCG65822.1
+ More
PCG65824.1 PCG65823.1 ODYU01006515 SOQ48427.1 KQ460466 KPJ14625.1 KQ459597 KPI95231.1 JTDY01000369 KOB77498.1 GEZM01014005 JAV92228.1 GEZM01014006 JAV92226.1 KQ971307 EFA10944.1 APGK01047275 KB741077 KB632432 ENN74318.1 ERL95545.1 GECU01010928 JAS96778.1 GECZ01004475 JAS65294.1 GECU01002492 JAT05215.1 GECZ01000946 JAS68823.1 GEDC01024731 JAS12567.1 ABLF02027949 ABLF02027951 ABLF02027953 ABLF02027954 GEDC01000082 JAS37216.1 CVRI01000074 CRL08238.1 GEBQ01012204 JAT27773.1 GECZ01006591 JAS63178.1 GBRD01009907 GDHC01015066 JAG55917.1 JAQ03563.1 GECZ01011151 JAS58618.1 GECU01004553 JAT03154.1 GEBQ01007305 JAT32672.1 KQ978350 KYM94716.1 GECZ01023618 JAS46151.1 GBHO01037798 JAG05806.1 GECU01013780 JAS93926.1 GEDC01030870 JAS06428.1 KQ983167 KYQ46931.1 GDHC01011438 JAQ07191.1 KK852708 KDR17984.1 KQ434819 KZC06920.1 GGMR01016248 MBY28867.1 KK107119 QOIP01000009 EZA58817.1 RLU19076.1 GGMR01007613 MBY20232.1 GEDC01006938 JAS30360.1 GL443213 EFN62425.1 GEBQ01020243 JAT19734.1 LJIG01016297 KRT81023.1 GEBQ01029219 JAT10758.1 GAMC01011594 JAB94961.1 UFQS01000680 UFQT01000680 SSX06160.1 SSX26516.1 CH902641 KPU74698.1 EDV33719.1 KPU74699.1 GL452536 EFN77097.1
PCG65824.1 PCG65823.1 ODYU01006515 SOQ48427.1 KQ460466 KPJ14625.1 KQ459597 KPI95231.1 JTDY01000369 KOB77498.1 GEZM01014005 JAV92228.1 GEZM01014006 JAV92226.1 KQ971307 EFA10944.1 APGK01047275 KB741077 KB632432 ENN74318.1 ERL95545.1 GECU01010928 JAS96778.1 GECZ01004475 JAS65294.1 GECU01002492 JAT05215.1 GECZ01000946 JAS68823.1 GEDC01024731 JAS12567.1 ABLF02027949 ABLF02027951 ABLF02027953 ABLF02027954 GEDC01000082 JAS37216.1 CVRI01000074 CRL08238.1 GEBQ01012204 JAT27773.1 GECZ01006591 JAS63178.1 GBRD01009907 GDHC01015066 JAG55917.1 JAQ03563.1 GECZ01011151 JAS58618.1 GECU01004553 JAT03154.1 GEBQ01007305 JAT32672.1 KQ978350 KYM94716.1 GECZ01023618 JAS46151.1 GBHO01037798 JAG05806.1 GECU01013780 JAS93926.1 GEDC01030870 JAS06428.1 KQ983167 KYQ46931.1 GDHC01011438 JAQ07191.1 KK852708 KDR17984.1 KQ434819 KZC06920.1 GGMR01016248 MBY28867.1 KK107119 QOIP01000009 EZA58817.1 RLU19076.1 GGMR01007613 MBY20232.1 GEDC01006938 JAS30360.1 GL443213 EFN62425.1 GEBQ01020243 JAT19734.1 LJIG01016297 KRT81023.1 GEBQ01029219 JAT10758.1 GAMC01011594 JAB94961.1 UFQS01000680 UFQT01000680 SSX06160.1 SSX26516.1 CH902641 KPU74698.1 EDV33719.1 KPU74699.1 GL452536 EFN77097.1
Proteomes
PRIDE
Interpro
ProteinModelPortal
H9JUY9
A0A2A4J1F4
A0A2A4J301
A0A2A4J340
A0A2A4J2T0
A0A2A4J351
+ More
A0A2H1W5W7 A0A0N0PCV7 A0A194PP70 A0A0L7LPP8 A0A1W4WJL8 A0A1W4WKK1 A0A1W4WJL3 A0A1Y1N493 A0A1Y1N7K5 D6W8F2 N6T9B3 A0A1B6JCB9 A0A1B6GS96 A0A1B6K198 A0A1B6H2C3 A0A1B6CGE6 X1WJN8 A0A1B6EH04 A0A1J1J9P0 A0A1B6LVR4 A0A1B6GL89 A0A0K8SRB8 A0A1B6G839 A0A1B6JW54 A0A1B6M9R2 A0A195C3F0 A0A1B6F7Q4 A0A0A9WEI4 A0A1B6J430 A0A1B6BYV1 A0A151WGI1 A0A146LGC7 A0A067RDG1 A0A154P719 A0A2S2PHH2 A0A026WS15 A0A2S2NT65 A0A1B6DXH0 E2AW03 A0A1W4W9I4 A0A1W4WKU2 A0A1B6L878 A0A0T6B0N5 A0A1B6KH13 W8BNL0 A0A336KYY0 K7J354 A0A0P8XII8 B3N0M1 E2C4L7
A0A2H1W5W7 A0A0N0PCV7 A0A194PP70 A0A0L7LPP8 A0A1W4WJL8 A0A1W4WKK1 A0A1W4WJL3 A0A1Y1N493 A0A1Y1N7K5 D6W8F2 N6T9B3 A0A1B6JCB9 A0A1B6GS96 A0A1B6K198 A0A1B6H2C3 A0A1B6CGE6 X1WJN8 A0A1B6EH04 A0A1J1J9P0 A0A1B6LVR4 A0A1B6GL89 A0A0K8SRB8 A0A1B6G839 A0A1B6JW54 A0A1B6M9R2 A0A195C3F0 A0A1B6F7Q4 A0A0A9WEI4 A0A1B6J430 A0A1B6BYV1 A0A151WGI1 A0A146LGC7 A0A067RDG1 A0A154P719 A0A2S2PHH2 A0A026WS15 A0A2S2NT65 A0A1B6DXH0 E2AW03 A0A1W4W9I4 A0A1W4WKU2 A0A1B6L878 A0A0T6B0N5 A0A1B6KH13 W8BNL0 A0A336KYY0 K7J354 A0A0P8XII8 B3N0M1 E2C4L7
Ontologies
GO
PANTHER
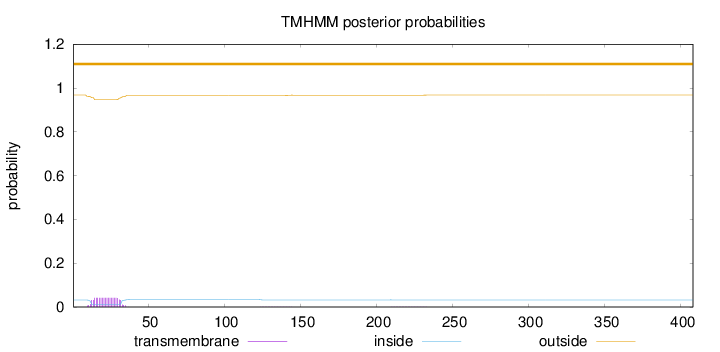
Topology
Subcellular location
Nucleus
Length:
408
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.885459999999999
Exp number, first 60 AAs:
0.84829
Total prob of N-in:
0.03200
outside
1 - 408
Population Genetic Test Statistics
Pi
275.244939
Theta
193.042952
Tajima's D
1.472027
CLR
0
CSRT
0.777261136943153
Interpretation
Uncertain
