Gene
KWMTBOMO15861
Pre Gene Modal
BGIBMGA013355
Annotation
PREDICTED:_potassium_voltage-gated_channel_protein_Shal_isoform_X3_[Amyelois_transitella]
Full name
Potassium voltage-gated channel protein Shal
Alternative Name
Shaker cognate l
Shal2
Shal2
Location in the cell
PlasmaMembrane Reliability : 4.577
Sequence
CDS
ATGGCGTCCGTCGCCGCCTGGCTGCCCTTCGCCAGAGCAGCAGCCATCGGGTGGGTCCCCATCGCGACCCATCCACTTCCGCCGCCGCCCGTGCCCAAGGACCGCCGCCGTGCCGAGGACGAGAAGCTCCTCATCAACGTCTCCGGACGACGCTTTGAGACTTGGCGGAACACTCTCGAGAAGTACCCGGACTCGCTGCTTGGTTCCTCAGAACGCGAATTCTTTTACGACGAAGACAGCCGAGAGTACTTTTTTGACAGAGACCCGGACATATTCAGGCATATTCTGAATTATTACCGCACTGGTAAACTCCACTACCCCAAACACGAGTGCCTCACGGGCTACGATGAGGAACTCGCGTTCTTCGGCATCCTTCCCGATGTCATCGGTGACTGTTGTTACGAAGACTACAGAGACAGGAAGAGAGAAAATGCCGAACGTCTTATGGATGACAAGCTTAGTGAAGCAGGCGACCAGAGTCTGCCTCAGCTCACGGACTTACGACAAAAAATGTGGAGGGCCTTCGAGAACCCCCACACGTCAACGGCTGCCCTCGTGTTTTACTATGTGACTGGATTTTTCATTGCCGTCTCCGTGATGGCTAACGTAGTGGAGACGGTACCGTGCGGACACCGACCTGGCCGGGCCGGGACCCTGCCGTGCGGCGAACGCTACAAGATAGTGTTCTTCTGCCTGGACACGGCGTGCGTGATGATCTTCACGGCGGAGTACCTGCTGCGACTGTTCGCGGCCCCGGACCGCTGCAAGTTCGTTCGCTCGGTGATGTCCATCATCGACGTGGTCGCGATCCTGCCGTACTACATCGGCCTGGGTATCACTGACAACGACGACGTCTCGGGTGCGTTTGTCACCCTCAGAGTGTTCCGAGTGTTTCGGATCTTCAAGTTCTCGCGGCACTCCCAAGGACTAAGGATCCTGGGCTACACGCTCAAGTCGTGCGCCAGCGAGCTCGGCTTCCTGGTGTTCTCGCTCGCGATGGCCATCATTATATTCGCGACAGTGATGTTCTACGCGGAGAAGAACGAGCAGGACACAAACTTCACGTCGATTCCGGCCGCCTTCTGGTACACCATCGTCACGATGACCACGCTCGGGTAA
Protein
MASVAAWLPFARAAAIGWVPIATHPLPPPPVPKDRRRAEDEKLLINVSGRRFETWRNTLEKYPDSLLGSSEREFFYDEDSREYFFDRDPDIFRHILNYYRTGKLHYPKHECLTGYDEELAFFGILPDVIGDCCYEDYRDRKRENAERLMDDKLSEAGDQSLPQLTDLRQKMWRAFENPHTSTAALVFYYVTGFFIAVSVMANVVETVPCGHRPGRAGTLPCGERYKIVFFCLDTACVMIFTAEYLLRLFAAPDRCKFVRSVMSIIDVVAILPYYIGLGITDNDDVSGAFVTLRVFRVFRIFKFSRHSQGLRILGYTLKSCASELGFLVFSLAMAIIIFATVMFYAEKNEQDTNFTSIPAAFWYTIVTMTTLG
Summary
Subunit
Heterotetramer of potassium channel proteins (By similarity). Interacts (via C-terminal dendritic targeting motif) with SIDL.
Miscellaneous
The segment S4 is probably the voltage-sensor and is characterized by a series of positively charged amino acids at every third position.
Similarity
Belongs to the potassium channel family.
Belongs to the potassium channel family. D (Shal) (TC 1.A.1.2) subfamily. Shal sub-subfamily.
Belongs to the potassium channel family. D (Shal) (TC 1.A.1.2) subfamily. Shal sub-subfamily.
Keywords
Alternative splicing
Cell projection
Complete proteome
Glycoprotein
Ion channel
Ion transport
Membrane
Potassium
Potassium channel
Potassium transport
Reference proteome
Transmembrane
Transmembrane helix
Transport
Voltage-gated channel
Feature
chain Potassium voltage-gated channel protein Shal
splice variant In isoform A.
splice variant In isoform A.
Uniprot
A0A194PR55
A0A212EN18
A0A0N1IHR9
A0A2A4J3B2
A0A2H1W5S3
A0A0L7LCB6
+ More
A0A3Q0IM34 A0A2R7WQX1 T1HW31 A0A2H8U287 A0A2S1ZNL5 B4J1C1 A0A0A1XDD8 A0A0A1XKY8 A0A1I8P3U8 A0A1I8P3Q6 A0A0K8UEX6 A0A034VF95 B4KZC6 A0A0L0C821 B4LCL8 A0A0Q9WKS9 A0A1B0FMG4 A0A0Q9XCX0 A0A1A9XLY8 A0A1A9UT12 A0A1B0B626 J9JVQ3 A0A1I8P3U0 A0A0A1XA98 A0A1A9Z0E7 A0A1A9X5N2 A0A0Q5U5J9 P17971 A0A1W4UT45 A0A0Q9WJQ5 A0A1I8MQI2 A0A0Q9XNA3 M9PFP1 A0A0P8YBQ8 A0A1W4URC2 A0A0Q5UHC6 P17971-1 A0A1W4UE14 A0A0Q9WUN0 B3M701 B4IIM4 B3NIF3 W8C343 A0A2J7PXU2 A0A067QL12 B4MXL6 A0A0M4EIW8 A0A0R3P720 K7IY22 E0VBM4 A0A3B0JV30 Q29DG1 B4H997 A0A1W4WRP6 A0A0R3P755 A0A154PM26 A0A195BA78 A0A2A3ED36 A0A195ETU0 A0A087ZTU2 V9IEP6 A0A195E528 A0A1L8DF39 A0A195D0I0 A0A151X609 A0A0L7RBH1 V9IF76 A0A0N0BDX4 A0A310SCK9 D6W6N1 A0A0C9QGR5 A0A026WNN5 A0A3L8DV17 A0A158NWY3 E9IQT6 N6TY00 A0A0T6AZF0 E2A858 A0A0J7KX79 A0A0J9RY24 A0A182R109 A0A182NJP2 A0A182L399 Q7QIS7 W5JNE5 A0A182Y273 A0A182RXN9 Q173M3 A0A084WJW3 A0A182J361 A0A182MFM1 A0A182P7Q5 A0A182VIS1 A0A182K028
A0A3Q0IM34 A0A2R7WQX1 T1HW31 A0A2H8U287 A0A2S1ZNL5 B4J1C1 A0A0A1XDD8 A0A0A1XKY8 A0A1I8P3U8 A0A1I8P3Q6 A0A0K8UEX6 A0A034VF95 B4KZC6 A0A0L0C821 B4LCL8 A0A0Q9WKS9 A0A1B0FMG4 A0A0Q9XCX0 A0A1A9XLY8 A0A1A9UT12 A0A1B0B626 J9JVQ3 A0A1I8P3U0 A0A0A1XA98 A0A1A9Z0E7 A0A1A9X5N2 A0A0Q5U5J9 P17971 A0A1W4UT45 A0A0Q9WJQ5 A0A1I8MQI2 A0A0Q9XNA3 M9PFP1 A0A0P8YBQ8 A0A1W4URC2 A0A0Q5UHC6 P17971-1 A0A1W4UE14 A0A0Q9WUN0 B3M701 B4IIM4 B3NIF3 W8C343 A0A2J7PXU2 A0A067QL12 B4MXL6 A0A0M4EIW8 A0A0R3P720 K7IY22 E0VBM4 A0A3B0JV30 Q29DG1 B4H997 A0A1W4WRP6 A0A0R3P755 A0A154PM26 A0A195BA78 A0A2A3ED36 A0A195ETU0 A0A087ZTU2 V9IEP6 A0A195E528 A0A1L8DF39 A0A195D0I0 A0A151X609 A0A0L7RBH1 V9IF76 A0A0N0BDX4 A0A310SCK9 D6W6N1 A0A0C9QGR5 A0A026WNN5 A0A3L8DV17 A0A158NWY3 E9IQT6 N6TY00 A0A0T6AZF0 E2A858 A0A0J7KX79 A0A0J9RY24 A0A182R109 A0A182NJP2 A0A182L399 Q7QIS7 W5JNE5 A0A182Y273 A0A182RXN9 Q173M3 A0A084WJW3 A0A182J361 A0A182MFM1 A0A182P7Q5 A0A182VIS1 A0A182K028
Pubmed
26354079
22118469
26227816
17994087
25830018
25348373
+ More
26108605 18057021 2333511 2336395 10731132 12537572 20550966 25315136 12537568 12537573 12537574 16110336 17569856 17569867 24495485 24845553 15632085 20075255 20566863 18362917 19820115 24508170 30249741 21347285 21282665 23537049 20798317 22936249 20966253 12364791 14747013 17210077 20920257 23761445 25244985 17510324 24438588
26108605 18057021 2333511 2336395 10731132 12537572 20550966 25315136 12537568 12537573 12537574 16110336 17569856 17569867 24495485 24845553 15632085 20075255 20566863 18362917 19820115 24508170 30249741 21347285 21282665 23537049 20798317 22936249 20966253 12364791 14747013 17210077 20920257 23761445 25244985 17510324 24438588
EMBL
KQ459597
KPI95229.1
AGBW02013760
OWR42869.1
KQ460466
KPJ14627.1
+ More
NWSH01003471 PCG66226.1 ODYU01006515 SOQ48431.1 JTDY01001805 KOB72836.1 KK855317 PTY21967.1 ACPB03001681 ACPB03001682 GFXV01007813 MBW19618.1 MF359242 AWK26945.1 CH916366 EDV97990.1 GBXI01005809 JAD08483.1 GBXI01002680 JAD11612.1 GDHF01027171 GDHF01012288 GDHF01003580 JAI25143.1 JAI40026.1 JAI48734.1 GAKP01017790 GAKP01017788 GAKP01017786 JAC41164.1 CH933809 EDW18952.1 JRES01000778 KNC28395.1 CH940647 EDW69881.1 KRF84625.1 KRF84627.1 KRF84629.1 KRF84631.1 KRF84632.1 KRF84630.1 CCAG010017555 KRG06431.1 JXJN01008949 ABLF02039740 ABLF02039755 ABLF02039756 ABLF02044019 GBXI01006471 JAD07821.1 CH954178 KQS44245.1 KQS44246.1 M32660 AE014296 BT024203 KRF84626.1 KRG06430.1 AGB94748.1 CH902618 KPU78821.1 KQS44243.1 KQS44244.1 KRF84628.1 EDV40866.1 CH480844 EDW49795.1 EDV52378.1 GAMC01010327 GAMC01010326 GAMC01010323 JAB96229.1 NEVH01020852 PNF21151.1 KK853224 KDR09686.1 CH963876 EDW76785.1 CP012525 ALC44765.1 CH379070 KRT08970.1 DS235033 EEB10780.1 OUUW01000002 SPP76601.1 EAL30453.2 CH479227 EDW35321.1 KRT08971.1 KQ434977 KZC12902.1 KQ976542 KYM81130.1 KZ288291 PBC29202.1 KQ981965 KYN31665.1 JR040403 AEY59117.1 KQ979608 KYN20290.1 GFDF01009114 JAV04970.1 KQ977004 KYN06423.1 KQ982482 KYQ55835.1 KQ414617 KOC68214.1 JR040402 AEY59116.1 KQ435851 KOX70822.1 KQ778189 OAD52013.1 KQ971307 EFA10968.1 GBYB01013763 JAG83530.1 KK107144 EZA57610.1 QOIP01000003 RLU24320.1 ADTU01028805 GL764900 EFZ17091.1 APGK01047179 KB741077 KB632384 ENN74185.1 ERL94320.1 LJIG01022461 KRT80456.1 GL437497 EFN70370.1 LBMM01002395 KMQ94879.1 CM002912 KMZ00564.1 AXCN02000434 AAAB01008807 EAA03996.2 ADMH02000859 ETN64838.1 CH477421 EAT41230.1 ATLV01024081 KE525348 KFB50507.1 AXCM01007912
NWSH01003471 PCG66226.1 ODYU01006515 SOQ48431.1 JTDY01001805 KOB72836.1 KK855317 PTY21967.1 ACPB03001681 ACPB03001682 GFXV01007813 MBW19618.1 MF359242 AWK26945.1 CH916366 EDV97990.1 GBXI01005809 JAD08483.1 GBXI01002680 JAD11612.1 GDHF01027171 GDHF01012288 GDHF01003580 JAI25143.1 JAI40026.1 JAI48734.1 GAKP01017790 GAKP01017788 GAKP01017786 JAC41164.1 CH933809 EDW18952.1 JRES01000778 KNC28395.1 CH940647 EDW69881.1 KRF84625.1 KRF84627.1 KRF84629.1 KRF84631.1 KRF84632.1 KRF84630.1 CCAG010017555 KRG06431.1 JXJN01008949 ABLF02039740 ABLF02039755 ABLF02039756 ABLF02044019 GBXI01006471 JAD07821.1 CH954178 KQS44245.1 KQS44246.1 M32660 AE014296 BT024203 KRF84626.1 KRG06430.1 AGB94748.1 CH902618 KPU78821.1 KQS44243.1 KQS44244.1 KRF84628.1 EDV40866.1 CH480844 EDW49795.1 EDV52378.1 GAMC01010327 GAMC01010326 GAMC01010323 JAB96229.1 NEVH01020852 PNF21151.1 KK853224 KDR09686.1 CH963876 EDW76785.1 CP012525 ALC44765.1 CH379070 KRT08970.1 DS235033 EEB10780.1 OUUW01000002 SPP76601.1 EAL30453.2 CH479227 EDW35321.1 KRT08971.1 KQ434977 KZC12902.1 KQ976542 KYM81130.1 KZ288291 PBC29202.1 KQ981965 KYN31665.1 JR040403 AEY59117.1 KQ979608 KYN20290.1 GFDF01009114 JAV04970.1 KQ977004 KYN06423.1 KQ982482 KYQ55835.1 KQ414617 KOC68214.1 JR040402 AEY59116.1 KQ435851 KOX70822.1 KQ778189 OAD52013.1 KQ971307 EFA10968.1 GBYB01013763 JAG83530.1 KK107144 EZA57610.1 QOIP01000003 RLU24320.1 ADTU01028805 GL764900 EFZ17091.1 APGK01047179 KB741077 KB632384 ENN74185.1 ERL94320.1 LJIG01022461 KRT80456.1 GL437497 EFN70370.1 LBMM01002395 KMQ94879.1 CM002912 KMZ00564.1 AXCN02000434 AAAB01008807 EAA03996.2 ADMH02000859 ETN64838.1 CH477421 EAT41230.1 ATLV01024081 KE525348 KFB50507.1 AXCM01007912
Proteomes
UP000053268
UP000007151
UP000053240
UP000218220
UP000037510
UP000079169
+ More
UP000015103 UP000001070 UP000095300 UP000009192 UP000037069 UP000008792 UP000092444 UP000092443 UP000078200 UP000092460 UP000007819 UP000092445 UP000091820 UP000008711 UP000000803 UP000192221 UP000095301 UP000007801 UP000001292 UP000235965 UP000027135 UP000007798 UP000092553 UP000001819 UP000002358 UP000009046 UP000268350 UP000008744 UP000192223 UP000076502 UP000078540 UP000242457 UP000078541 UP000005203 UP000078492 UP000078542 UP000075809 UP000053825 UP000053105 UP000007266 UP000053097 UP000279307 UP000005205 UP000019118 UP000030742 UP000000311 UP000036403 UP000075886 UP000075884 UP000075882 UP000007062 UP000000673 UP000076408 UP000075900 UP000008820 UP000030765 UP000075880 UP000075883 UP000075885 UP000075903 UP000075881
UP000015103 UP000001070 UP000095300 UP000009192 UP000037069 UP000008792 UP000092444 UP000092443 UP000078200 UP000092460 UP000007819 UP000092445 UP000091820 UP000008711 UP000000803 UP000192221 UP000095301 UP000007801 UP000001292 UP000235965 UP000027135 UP000007798 UP000092553 UP000001819 UP000002358 UP000009046 UP000268350 UP000008744 UP000192223 UP000076502 UP000078540 UP000242457 UP000078541 UP000005203 UP000078492 UP000078542 UP000075809 UP000053825 UP000053105 UP000007266 UP000053097 UP000279307 UP000005205 UP000019118 UP000030742 UP000000311 UP000036403 UP000075886 UP000075884 UP000075882 UP000007062 UP000000673 UP000076408 UP000075900 UP000008820 UP000030765 UP000075880 UP000075883 UP000075885 UP000075903 UP000075881
Interpro
SUPFAM
SSF54695
SSF54695
Gene 3D
ProteinModelPortal
A0A194PR55
A0A212EN18
A0A0N1IHR9
A0A2A4J3B2
A0A2H1W5S3
A0A0L7LCB6
+ More
A0A3Q0IM34 A0A2R7WQX1 T1HW31 A0A2H8U287 A0A2S1ZNL5 B4J1C1 A0A0A1XDD8 A0A0A1XKY8 A0A1I8P3U8 A0A1I8P3Q6 A0A0K8UEX6 A0A034VF95 B4KZC6 A0A0L0C821 B4LCL8 A0A0Q9WKS9 A0A1B0FMG4 A0A0Q9XCX0 A0A1A9XLY8 A0A1A9UT12 A0A1B0B626 J9JVQ3 A0A1I8P3U0 A0A0A1XA98 A0A1A9Z0E7 A0A1A9X5N2 A0A0Q5U5J9 P17971 A0A1W4UT45 A0A0Q9WJQ5 A0A1I8MQI2 A0A0Q9XNA3 M9PFP1 A0A0P8YBQ8 A0A1W4URC2 A0A0Q5UHC6 P17971-1 A0A1W4UE14 A0A0Q9WUN0 B3M701 B4IIM4 B3NIF3 W8C343 A0A2J7PXU2 A0A067QL12 B4MXL6 A0A0M4EIW8 A0A0R3P720 K7IY22 E0VBM4 A0A3B0JV30 Q29DG1 B4H997 A0A1W4WRP6 A0A0R3P755 A0A154PM26 A0A195BA78 A0A2A3ED36 A0A195ETU0 A0A087ZTU2 V9IEP6 A0A195E528 A0A1L8DF39 A0A195D0I0 A0A151X609 A0A0L7RBH1 V9IF76 A0A0N0BDX4 A0A310SCK9 D6W6N1 A0A0C9QGR5 A0A026WNN5 A0A3L8DV17 A0A158NWY3 E9IQT6 N6TY00 A0A0T6AZF0 E2A858 A0A0J7KX79 A0A0J9RY24 A0A182R109 A0A182NJP2 A0A182L399 Q7QIS7 W5JNE5 A0A182Y273 A0A182RXN9 Q173M3 A0A084WJW3 A0A182J361 A0A182MFM1 A0A182P7Q5 A0A182VIS1 A0A182K028
A0A3Q0IM34 A0A2R7WQX1 T1HW31 A0A2H8U287 A0A2S1ZNL5 B4J1C1 A0A0A1XDD8 A0A0A1XKY8 A0A1I8P3U8 A0A1I8P3Q6 A0A0K8UEX6 A0A034VF95 B4KZC6 A0A0L0C821 B4LCL8 A0A0Q9WKS9 A0A1B0FMG4 A0A0Q9XCX0 A0A1A9XLY8 A0A1A9UT12 A0A1B0B626 J9JVQ3 A0A1I8P3U0 A0A0A1XA98 A0A1A9Z0E7 A0A1A9X5N2 A0A0Q5U5J9 P17971 A0A1W4UT45 A0A0Q9WJQ5 A0A1I8MQI2 A0A0Q9XNA3 M9PFP1 A0A0P8YBQ8 A0A1W4URC2 A0A0Q5UHC6 P17971-1 A0A1W4UE14 A0A0Q9WUN0 B3M701 B4IIM4 B3NIF3 W8C343 A0A2J7PXU2 A0A067QL12 B4MXL6 A0A0M4EIW8 A0A0R3P720 K7IY22 E0VBM4 A0A3B0JV30 Q29DG1 B4H997 A0A1W4WRP6 A0A0R3P755 A0A154PM26 A0A195BA78 A0A2A3ED36 A0A195ETU0 A0A087ZTU2 V9IEP6 A0A195E528 A0A1L8DF39 A0A195D0I0 A0A151X609 A0A0L7RBH1 V9IF76 A0A0N0BDX4 A0A310SCK9 D6W6N1 A0A0C9QGR5 A0A026WNN5 A0A3L8DV17 A0A158NWY3 E9IQT6 N6TY00 A0A0T6AZF0 E2A858 A0A0J7KX79 A0A0J9RY24 A0A182R109 A0A182NJP2 A0A182L399 Q7QIS7 W5JNE5 A0A182Y273 A0A182RXN9 Q173M3 A0A084WJW3 A0A182J361 A0A182MFM1 A0A182P7Q5 A0A182VIS1 A0A182K028
PDB
2I2R
E-value=1.70736e-58,
Score=572
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
Cell projection
Dendrite
Perikaryon
Cell projection
Dendrite
Perikaryon
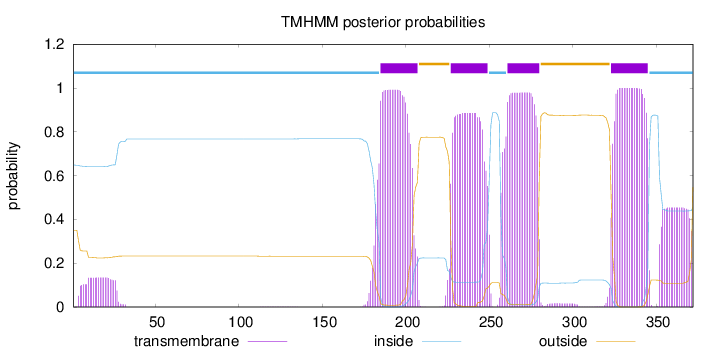
Length:
372
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
96.77039
Exp number, first 60 AAs:
2.94086
Total prob of N-in:
0.64916
inside
1 - 184
TMhelix
185 - 207
outside
208 - 226
TMhelix
227 - 249
inside
250 - 260
TMhelix
261 - 280
outside
281 - 322
TMhelix
323 - 345
inside
346 - 372
Population Genetic Test Statistics
Pi
324.937729
Theta
237.440582
Tajima's D
1.133166
CLR
0
CSRT
0.696565171741413
Interpretation
Uncertain
