Gene
KWMTBOMO15850
Pre Gene Modal
BGIBMGA000063
Annotation
PREDICTED:_serine/threonine-protein_kinase_pelle-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.458 Nuclear Reliability : 1.771
Sequence
CDS
ATGTACATTTACGAATTACCACCCGAGATTAACAAAGAACTATGTCGACTTTTGGACAACTTCGATTACTGGGAGGAGCTGGCCGGCAATTACATGATGTACACAGCGATGGACGTCATCGAAATAAAAGAGAAGGCTCGTCATCAGGGCATATCCCCAACGGAATACCTCCTGGAGTGCTGGGGGCAGAAGAACCATCGAATTGAGGAACTGTTTGTGTTGCTGTACCGCATGAAGCACATTCCTGCCATGAGGCGGCTAACCGGCGTCGTGCACGAGAGGTTCCATAGACTCCTACACAAGCTGGAGGCGGGGAACGGACAGGATGGAAACAACGCGCGCACCGGCGGCGCGCAGAGGGCCACCACCAGCAGCGACTCCGTCCCGCTCCCGGTCATACTCTTTGATCAGGGGATCAGGCAGGCTCCCTCGAGCAGCGGACTCAACGGGCGCTCTCCCTCCGCGGACACGAGCTCGGCCACCAGCACCAGCACCGACACCTCGTCCGCGCAGTACGACAAGTACGATGAGATCCTGCGCAAGGTGTCCGCCATCCCGAAGATGAGCTACGAGGAGCTGCGGCTCGCCACGGACGACTGGAGCGAGCGCAACCTGCTCGGCCGCGGCGGCTTCGGACAGGTCTACAAGGGGGAGTGGAAGCTGCTGGGCGTGGCGGTGAAGAGGCTGCGCGACAATGAGGGCCGCGACCAAGAGCTCGTGCGGGAGATGTGCCTCAACCAGTACCGCCACGACAACATCCTGCCCCTCTACGGATACAGCCTGGGAGGGCCCGAGGCGTGCCTGGTGTACCAGCTGATGGCGGGCGGCTCGCTGGAGCAGCGGCTGCGGCCCAAGGACGGCGCCGCGCCGCTCTCCTGGGCGCAGCGCTGCAGGATCATGTGCGGGGTGGCCAGAGGGCTGCAGTTCCTGCACACGATGACCGGCAGCGCGCTCATCCACGGGGACATCAAGCCGGCTAACATCTTGCTGGACCAGTGCCTCGAGCCCAAGATCGGAGACTTCGGCCTCGCGCGGAAGGGGCCCTACGGAGACGAACGGACGCATCTGAAGGTCTCACGAGTGTACGGGACGAGGCCCTACTTGCCGGACGAGTACCTGCAGTCGTGCGTGCTGTCGCCGGCCGTCGACGTGTACAGCTTCGGCGTGGTGGTGGCCGAGACCAGCACGGGGCTGCCGGCCTGGGACCGCTCCCGCGCGCGGCCCCTGCTGGCGCACCAGCTGCACTCGTGGCACGCCGCGCGGCACGACCTGGGCGCGCTGCACGACCCGCGCCTGCCGCCGCACCCCGCGCTGTGCCGCGCGCTGCTGGAGCTGGCGCTGCACTGCACGCAGCCGCCGCGCGAGGCGCGCCCGCCCATGCTGCTCGTGTACCGCCGCCTGCACGCGCTCGCCGCCTGCTAG
Protein
MYIYELPPEINKELCRLLDNFDYWEELAGNYMMYTAMDVIEIKEKARHQGISPTEYLLECWGQKNHRIEELFVLLYRMKHIPAMRRLTGVVHERFHRLLHKLEAGNGQDGNNARTGGAQRATTSSDSVPLPVILFDQGIRQAPSSSGLNGRSPSADTSSATSTSTDTSSAQYDKYDEILRKVSAIPKMSYEELRLATDDWSERNLLGRGGFGQVYKGEWKLLGVAVKRLRDNEGRDQELVREMCLNQYRHDNILPLYGYSLGGPEACLVYQLMAGGSLEQRLRPKDGAAPLSWAQRCRIMCGVARGLQFLHTMTGSALIHGDIKPANILLDQCLEPKIGDFGLARKGPYGDERTHLKVSRVYGTRPYLPDEYLQSCVLSPAVDVYSFGVVVAETSTGLPAWDRSRARPLLAHQLHSWHAARHDLGALHDPRLPPHPALCRALLELALHCTQPPREARPPMLLVYRRLHALAAC
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
H9IS37
A0A2A4KAG9
A0A3G2LU65
A0A194QJI6
A0A3S2M6U4
A0A212ELF2
+ More
A0A2H1V8B7 A0A2A4K933 A0A1L8DKU7 A0A1Y1K6D8 D2A4G0 E9IUH3 F4WJM5 E2AWE1 A0A151WH44 A0A158NND5 A0A195FWM4 A0A195E4Q7 A0A0L7QWX7 T1HNR1 E2BXL3 A0A151I695 A0A0A9Y0R5 A0A0P4VTW0 A0A0V0GA25 A0A023F0Z6 A0A1A9WJS4 A0A2S2RB22 A0A0J7K8M2 A0A224XDE4 A0A088AD23 A0A154PEP2 A0A2A3ELR7 A0A026X418 A0A2H8TMG5 A0A0M8ZS40 J9KAE6 A0A1A9XPX6 A0A0C9R5K5 A0A0P4ZA62 A0A0P5VR50 A0A0P4ZC16 A0A232EJJ2 A0A0P5T3L4 A0A0P5YQB5 A0A0P5DFL4 A0A0P6FSK5 K7J9T5 A0A0P5DYD1 A0A1B0BCQ1 A0A0P5DD66 A0A1B0FAS5 A0A1A9ZE77 A0A1A9VCY9 A0A0K8TNV8 A0A0P5QNH9 A0A0P5ETK9 A0A0M4F6V2 A0A0A9WJC8 N6TH67 U4US38 A0A182JUP9 A0A0P5R2I6 A0A226DFL9 A0A0P5C499 A0A0P5LDN3 A0A182NDW5 A0A0D3QDC6 Q7QD54 E0VYT1 A0A182WRQ6 A0A182TTZ0 A0A0D3QE27 A0A0D3QDC2 A0A0D3QDF3 A0A182HU22 A0A0D3QDP5 A0A0D3QDE8 A0A1J1HUD4 A0A0D3QE31 A0A0D3QDD5 A0A0D3QDY5 A0A0D3QE29 A0A0D3QDQ0 A0A0D3QDD1 A0A182P6N4 A0A0P4XED2 A0A182VYJ6 A0A182RNB0 A0A182LW73 A0A0L0CKI7 A0A0P5B6I8 A0A182SNV8 A0A2M4AN68 A0A0K8SHY7 A0A2M3ZJ21 A0A182XWC9 A0A182IPR1
A0A2H1V8B7 A0A2A4K933 A0A1L8DKU7 A0A1Y1K6D8 D2A4G0 E9IUH3 F4WJM5 E2AWE1 A0A151WH44 A0A158NND5 A0A195FWM4 A0A195E4Q7 A0A0L7QWX7 T1HNR1 E2BXL3 A0A151I695 A0A0A9Y0R5 A0A0P4VTW0 A0A0V0GA25 A0A023F0Z6 A0A1A9WJS4 A0A2S2RB22 A0A0J7K8M2 A0A224XDE4 A0A088AD23 A0A154PEP2 A0A2A3ELR7 A0A026X418 A0A2H8TMG5 A0A0M8ZS40 J9KAE6 A0A1A9XPX6 A0A0C9R5K5 A0A0P4ZA62 A0A0P5VR50 A0A0P4ZC16 A0A232EJJ2 A0A0P5T3L4 A0A0P5YQB5 A0A0P5DFL4 A0A0P6FSK5 K7J9T5 A0A0P5DYD1 A0A1B0BCQ1 A0A0P5DD66 A0A1B0FAS5 A0A1A9ZE77 A0A1A9VCY9 A0A0K8TNV8 A0A0P5QNH9 A0A0P5ETK9 A0A0M4F6V2 A0A0A9WJC8 N6TH67 U4US38 A0A182JUP9 A0A0P5R2I6 A0A226DFL9 A0A0P5C499 A0A0P5LDN3 A0A182NDW5 A0A0D3QDC6 Q7QD54 E0VYT1 A0A182WRQ6 A0A182TTZ0 A0A0D3QE27 A0A0D3QDC2 A0A0D3QDF3 A0A182HU22 A0A0D3QDP5 A0A0D3QDE8 A0A1J1HUD4 A0A0D3QE31 A0A0D3QDD5 A0A0D3QDY5 A0A0D3QE29 A0A0D3QDQ0 A0A0D3QDD1 A0A182P6N4 A0A0P4XED2 A0A182VYJ6 A0A182RNB0 A0A182LW73 A0A0L0CKI7 A0A0P5B6I8 A0A182SNV8 A0A2M4AN68 A0A0K8SHY7 A0A2M3ZJ21 A0A182XWC9 A0A182IPR1
Pubmed
EMBL
BABH01035689
BABH01035690
NWSH01000019
PCG80730.1
MG837006
AYN77138.1
+ More
KQ459144 KPJ03611.1 RSAL01000019 RVE52796.1 AGBW02014080 OWR42299.1 ODYU01001184 SOQ37051.1 PCG80731.1 GFDF01006998 JAV07086.1 GEZM01091300 JAV56943.1 KQ971344 EFA05720.1 GL765974 EFZ15781.1 GL888186 EGI65517.1 GL443286 EFN62241.1 KQ983136 KYQ47150.1 ADTU01021232 KQ981208 KYN44712.1 KQ979685 KYN19839.1 KQ414706 KOC63071.1 ACPB03016618 GL451265 EFN79582.1 KQ978495 KYM93626.1 GBHO01035703 GBHO01032216 GBHO01018891 JAG07901.1 JAG11388.1 JAG24713.1 GDKW01000516 JAI56079.1 GECL01001874 JAP04250.1 GBBI01003525 JAC15187.1 GGMS01017667 MBY86870.1 LBMM01011911 KMQ86556.1 GFTR01007422 JAW09004.1 KQ434889 KZC10301.1 KZ288220 PBC32146.1 KK107031 EZA62099.1 GFXV01003047 MBW14852.1 KQ435878 KOX69995.1 ABLF02025450 ABLF02025452 GBYB01003215 JAG72982.1 GDIP01216130 JAJ07272.1 GDIP01111040 JAL92674.1 GDIP01216131 JAJ07271.1 NNAY01004026 OXU18478.1 GDIP01135979 LRGB01002860 JAL67735.1 KZS05836.1 GDIP01054705 JAM49010.1 GDIP01160525 JAJ62877.1 GDIQ01043663 JAN51074.1 AAZX01005976 GDIP01153842 JAJ69560.1 JXJN01012081 GDIP01159339 JAJ64063.1 CCAG010007158 GDAI01001770 JAI15833.1 GDIQ01112333 JAL39393.1 GDIQ01265217 JAJ86507.1 CP012526 ALC47495.1 GBHO01035700 JAG07904.1 APGK01038380 KB740954 ENN77083.1 KB632338 ERL92890.1 GDIQ01126480 JAL25246.1 LNIX01000022 OXA43371.1 GDIP01191420 JAJ31982.1 GDIQ01196450 JAK55275.1 KP274740 KP274743 KP274746 KP274747 KP274749 AJC98906.1 AJC98909.1 AJC98912.1 AJC98913.1 AJC98915.1 AAAB01008859 EAA07539.4 DS235847 EEB18537.1 KP274748 AJC98914.1 KP274742 AJC98908.1 KP274760 AJC98926.1 APCN01000602 KP274741 KP274750 KP274756 AJC98907.1 AJC98916.1 AJC98922.1 KP274755 AJC98921.1 CVRI01000021 CRK91680.1 KP274758 AJC98924.1 KP274757 AJC98923.1 KP274754 AJC98920.1 KP274753 AJC98919.1 KP274745 KP274751 KP274759 AJC98911.1 AJC98917.1 AJC98925.1 KP274744 KP274752 AJC98910.1 AJC98918.1 GDIP01243007 JAI80394.1 AXCM01004068 JRES01000266 KNC32781.1 GDIP01188821 JAJ34581.1 GGFK01008908 MBW42229.1 GBRD01012898 GBRD01008431 GBRD01003361 GBRD01001564 GBRD01001561 JAG52928.1 GGFM01007714 MBW28465.1
KQ459144 KPJ03611.1 RSAL01000019 RVE52796.1 AGBW02014080 OWR42299.1 ODYU01001184 SOQ37051.1 PCG80731.1 GFDF01006998 JAV07086.1 GEZM01091300 JAV56943.1 KQ971344 EFA05720.1 GL765974 EFZ15781.1 GL888186 EGI65517.1 GL443286 EFN62241.1 KQ983136 KYQ47150.1 ADTU01021232 KQ981208 KYN44712.1 KQ979685 KYN19839.1 KQ414706 KOC63071.1 ACPB03016618 GL451265 EFN79582.1 KQ978495 KYM93626.1 GBHO01035703 GBHO01032216 GBHO01018891 JAG07901.1 JAG11388.1 JAG24713.1 GDKW01000516 JAI56079.1 GECL01001874 JAP04250.1 GBBI01003525 JAC15187.1 GGMS01017667 MBY86870.1 LBMM01011911 KMQ86556.1 GFTR01007422 JAW09004.1 KQ434889 KZC10301.1 KZ288220 PBC32146.1 KK107031 EZA62099.1 GFXV01003047 MBW14852.1 KQ435878 KOX69995.1 ABLF02025450 ABLF02025452 GBYB01003215 JAG72982.1 GDIP01216130 JAJ07272.1 GDIP01111040 JAL92674.1 GDIP01216131 JAJ07271.1 NNAY01004026 OXU18478.1 GDIP01135979 LRGB01002860 JAL67735.1 KZS05836.1 GDIP01054705 JAM49010.1 GDIP01160525 JAJ62877.1 GDIQ01043663 JAN51074.1 AAZX01005976 GDIP01153842 JAJ69560.1 JXJN01012081 GDIP01159339 JAJ64063.1 CCAG010007158 GDAI01001770 JAI15833.1 GDIQ01112333 JAL39393.1 GDIQ01265217 JAJ86507.1 CP012526 ALC47495.1 GBHO01035700 JAG07904.1 APGK01038380 KB740954 ENN77083.1 KB632338 ERL92890.1 GDIQ01126480 JAL25246.1 LNIX01000022 OXA43371.1 GDIP01191420 JAJ31982.1 GDIQ01196450 JAK55275.1 KP274740 KP274743 KP274746 KP274747 KP274749 AJC98906.1 AJC98909.1 AJC98912.1 AJC98913.1 AJC98915.1 AAAB01008859 EAA07539.4 DS235847 EEB18537.1 KP274748 AJC98914.1 KP274742 AJC98908.1 KP274760 AJC98926.1 APCN01000602 KP274741 KP274750 KP274756 AJC98907.1 AJC98916.1 AJC98922.1 KP274755 AJC98921.1 CVRI01000021 CRK91680.1 KP274758 AJC98924.1 KP274757 AJC98923.1 KP274754 AJC98920.1 KP274753 AJC98919.1 KP274745 KP274751 KP274759 AJC98911.1 AJC98917.1 AJC98925.1 KP274744 KP274752 AJC98910.1 AJC98918.1 GDIP01243007 JAI80394.1 AXCM01004068 JRES01000266 KNC32781.1 GDIP01188821 JAJ34581.1 GGFK01008908 MBW42229.1 GBRD01012898 GBRD01008431 GBRD01003361 GBRD01001564 GBRD01001561 JAG52928.1 GGFM01007714 MBW28465.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000007151
UP000007266
+ More
UP000007755 UP000000311 UP000075809 UP000005205 UP000078541 UP000078492 UP000053825 UP000015103 UP000008237 UP000078542 UP000091820 UP000036403 UP000005203 UP000076502 UP000242457 UP000053097 UP000053105 UP000007819 UP000092443 UP000215335 UP000076858 UP000002358 UP000092460 UP000092444 UP000092445 UP000078200 UP000092553 UP000019118 UP000030742 UP000075881 UP000198287 UP000075884 UP000007062 UP000009046 UP000076407 UP000075902 UP000075903 UP000075840 UP000183832 UP000075885 UP000075920 UP000075900 UP000075883 UP000037069 UP000075901 UP000076408 UP000075880
UP000007755 UP000000311 UP000075809 UP000005205 UP000078541 UP000078492 UP000053825 UP000015103 UP000008237 UP000078542 UP000091820 UP000036403 UP000005203 UP000076502 UP000242457 UP000053097 UP000053105 UP000007819 UP000092443 UP000215335 UP000076858 UP000002358 UP000092460 UP000092444 UP000092445 UP000078200 UP000092553 UP000019118 UP000030742 UP000075881 UP000198287 UP000075884 UP000007062 UP000009046 UP000076407 UP000075902 UP000075903 UP000075840 UP000183832 UP000075885 UP000075920 UP000075900 UP000075883 UP000037069 UP000075901 UP000076408 UP000075880
PRIDE
Interpro
IPR001245
Ser-Thr/Tyr_kinase_cat_dom
+ More
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR000488 Death_domain
IPR011029 DEATH-like_dom_sf
IPR000719 Prot_kinase_dom
IPR037924 Pelle_death
IPR017853 Glycoside_hydrolase_SF
IPR001360 Glyco_hydro_1
IPR004344 TTL/TTLL_fam
IPR013815 ATP_grasp_subdomain_1
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR000488 Death_domain
IPR011029 DEATH-like_dom_sf
IPR000719 Prot_kinase_dom
IPR037924 Pelle_death
IPR017853 Glycoside_hydrolase_SF
IPR001360 Glyco_hydro_1
IPR004344 TTL/TTLL_fam
IPR013815 ATP_grasp_subdomain_1
Gene 3D
ProteinModelPortal
H9IS37
A0A2A4KAG9
A0A3G2LU65
A0A194QJI6
A0A3S2M6U4
A0A212ELF2
+ More
A0A2H1V8B7 A0A2A4K933 A0A1L8DKU7 A0A1Y1K6D8 D2A4G0 E9IUH3 F4WJM5 E2AWE1 A0A151WH44 A0A158NND5 A0A195FWM4 A0A195E4Q7 A0A0L7QWX7 T1HNR1 E2BXL3 A0A151I695 A0A0A9Y0R5 A0A0P4VTW0 A0A0V0GA25 A0A023F0Z6 A0A1A9WJS4 A0A2S2RB22 A0A0J7K8M2 A0A224XDE4 A0A088AD23 A0A154PEP2 A0A2A3ELR7 A0A026X418 A0A2H8TMG5 A0A0M8ZS40 J9KAE6 A0A1A9XPX6 A0A0C9R5K5 A0A0P4ZA62 A0A0P5VR50 A0A0P4ZC16 A0A232EJJ2 A0A0P5T3L4 A0A0P5YQB5 A0A0P5DFL4 A0A0P6FSK5 K7J9T5 A0A0P5DYD1 A0A1B0BCQ1 A0A0P5DD66 A0A1B0FAS5 A0A1A9ZE77 A0A1A9VCY9 A0A0K8TNV8 A0A0P5QNH9 A0A0P5ETK9 A0A0M4F6V2 A0A0A9WJC8 N6TH67 U4US38 A0A182JUP9 A0A0P5R2I6 A0A226DFL9 A0A0P5C499 A0A0P5LDN3 A0A182NDW5 A0A0D3QDC6 Q7QD54 E0VYT1 A0A182WRQ6 A0A182TTZ0 A0A0D3QE27 A0A0D3QDC2 A0A0D3QDF3 A0A182HU22 A0A0D3QDP5 A0A0D3QDE8 A0A1J1HUD4 A0A0D3QE31 A0A0D3QDD5 A0A0D3QDY5 A0A0D3QE29 A0A0D3QDQ0 A0A0D3QDD1 A0A182P6N4 A0A0P4XED2 A0A182VYJ6 A0A182RNB0 A0A182LW73 A0A0L0CKI7 A0A0P5B6I8 A0A182SNV8 A0A2M4AN68 A0A0K8SHY7 A0A2M3ZJ21 A0A182XWC9 A0A182IPR1
A0A2H1V8B7 A0A2A4K933 A0A1L8DKU7 A0A1Y1K6D8 D2A4G0 E9IUH3 F4WJM5 E2AWE1 A0A151WH44 A0A158NND5 A0A195FWM4 A0A195E4Q7 A0A0L7QWX7 T1HNR1 E2BXL3 A0A151I695 A0A0A9Y0R5 A0A0P4VTW0 A0A0V0GA25 A0A023F0Z6 A0A1A9WJS4 A0A2S2RB22 A0A0J7K8M2 A0A224XDE4 A0A088AD23 A0A154PEP2 A0A2A3ELR7 A0A026X418 A0A2H8TMG5 A0A0M8ZS40 J9KAE6 A0A1A9XPX6 A0A0C9R5K5 A0A0P4ZA62 A0A0P5VR50 A0A0P4ZC16 A0A232EJJ2 A0A0P5T3L4 A0A0P5YQB5 A0A0P5DFL4 A0A0P6FSK5 K7J9T5 A0A0P5DYD1 A0A1B0BCQ1 A0A0P5DD66 A0A1B0FAS5 A0A1A9ZE77 A0A1A9VCY9 A0A0K8TNV8 A0A0P5QNH9 A0A0P5ETK9 A0A0M4F6V2 A0A0A9WJC8 N6TH67 U4US38 A0A182JUP9 A0A0P5R2I6 A0A226DFL9 A0A0P5C499 A0A0P5LDN3 A0A182NDW5 A0A0D3QDC6 Q7QD54 E0VYT1 A0A182WRQ6 A0A182TTZ0 A0A0D3QE27 A0A0D3QDC2 A0A0D3QDF3 A0A182HU22 A0A0D3QDP5 A0A0D3QDE8 A0A1J1HUD4 A0A0D3QE31 A0A0D3QDD5 A0A0D3QDY5 A0A0D3QE29 A0A0D3QDQ0 A0A0D3QDD1 A0A182P6N4 A0A0P4XED2 A0A182VYJ6 A0A182RNB0 A0A182LW73 A0A0L0CKI7 A0A0P5B6I8 A0A182SNV8 A0A2M4AN68 A0A0K8SHY7 A0A2M3ZJ21 A0A182XWC9 A0A182IPR1
PDB
5UIU
E-value=2.7563e-38,
Score=399
Ontologies
GO
PANTHER
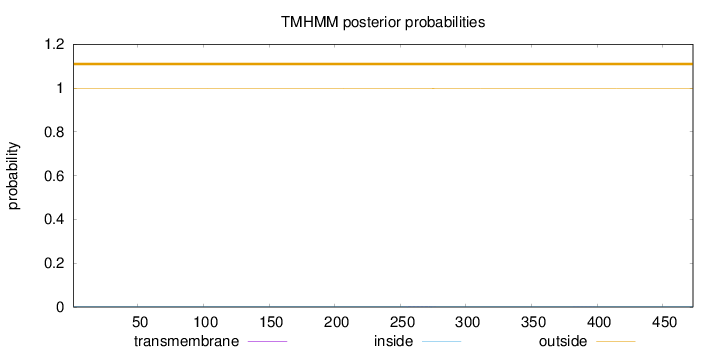
Topology
Length:
473
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0341
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.00185
outside
1 - 473
Population Genetic Test Statistics
Pi
154.363995
Theta
165.098459
Tajima's D
-0.639379
CLR
0.218693
CSRT
0.210089495525224
Interpretation
Uncertain
