Gene
KWMTBOMO15847 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000002
Annotation
PREDICTED:_uncharacterized_protein_LOC101745378_[Bombyx_mori]
Full name
MICOS complex subunit MIC60
Alternative Name
Mitofilin
Location in the cell
Cytoplasmic Reliability : 1.282 Mitochondrial Reliability : 1.395
Sequence
CDS
ATGTACAAATTTACGAATCATTTATCGAATGCTAAATTAGTTTTGGTAAGAAGACACATTGATGGGTCCTCAGCAGCACTGATAGTACTTCGTCAAAACTCATACTCCCAGGATCATGGTACACGACAGCAACCCCGCAAGTCACGCAAGCTCTTCTGGAGCACTGTCGGAGCGACTGTACTTACTGGAGCTGCTGTTGTTTATGCCAAACACAGTCCAGAGGCCCGAAACTGGTTAGAGTCAAATGTCCCTTGGGCCAATGACCTGGTAGCTCTCGTTTACCAGGAGAAGTCTTCACCGTGGAAGTTTACAGTGGACCGTTTCAACCAGGCCACCACGTCAATCAGCCACTTTGTGTTTGGAAAGGAAGGTGTTACGCCATTAGAGATAAAGTCGAGGTCACAGCAGGACATAGATGCAGATGCAGCAAAGAAGGCAACCAGCAAAGACTTCCAATTGCCTCCGCCCTCCCTTGAGCCCCTCTACGTGGAGGAGAAGAAGCTTGGAACACCGGATGTAGACACACCAGCTACGGTGGTGCAGACGGAGCGCTGCGAGCCCGCGCGCGCCCCGGCCGCCGCCGTGCCCGCCGAGCTGGTGCAGCTGGAGCGCGACATGATGGACAACACCAAGCTGGCCCTCGACAGCTACAAGCGCGCCTCCCAGCACTGCAAGCGCTACAACGACGCGCTCTACAAGATAGTGGAGTCCACCGTGGAGGAGCTGGACAAGAGGCACTTCTCGGCGCTGCAGGGCGCGCAGAGCGAGCGGGACGCGGCGCACGAGCAGGCCGCGGAGGCCGCCGCTCGCGCCAAGAGCGCCATCCAGCGGGTGGAGAGCGCGCTGGAGGCCGGGGCGCCGGTGCCGCCGCCCGCGGCCGCCGCCACCCGGCGCCACGCCGCCGCCTTCGGGGAGCAGCTGGCGGCCGCGGAGGCCGAGTACTGCCGGTGCCGGGACGAGGCGCTGCTGGCCGACCGCTACTGGGACAAGGTGGAGGCGGCCCGGTCTGCGTTCCGGGCGGAGCTGGCGGCGCTGTTCCCGGGCGCCGACCTCAGCGCGCGCTCCCTGCCCTCCGCGCACGTCGACCTGCTGCTCGTCTACACGCTCAAACAGATCCAGTTCCTGCAAAACCAGCTGGCGGAGCTGCAGACCGTGCGGGAGCAGAAGATCAACCGCGCCATTGAGTGTCACGACGAGAAGGCGCTCATCGAAGCTAAAGTGGAGGACCTAATCAAAGCGGAACGCGTCGAGAGGGAGAAGGAATTCCTCAAGAGGAGCCTGGCGCTGCAGGCCGAGGCCCACAGGAGCCTCCGGGAGCAGCTCAAGAAGCAGTTCGAGATACAGCAGGAGGTCCTGCAGGAGAAGGTCGCCAGCAAGGAGAAAGAGGTGATGTCCCGCCTGGCGCGCGCGCAGTCCGAGCGCCTGGAGCACGAGCGCGCGCAGCACAAGCGCGAGCTGGGCGCCGCCGCCGCGCGCCTCGCCGCCGTGCAGCACGCGCTCAAACAGCGGGCGGGCGCGGAGGAGGAGGCGCGGCGCAGCGCGGCGCTGTGGGCGGCGGCGGGCGGCGTGCTGGCCGCGCTGTCCGCGCCGCGCCGGGCGCCGCTGCGGGACCACGTGGCCGCCGTGCGCAGCGCCGGTAAGGACGACAAATTGGTGCAGAGCATCCTGCAGAGCATCTCGCGAGACGCGCTGGAGCACGGCGTGGCCACCGAGCAGGAGCTCCGGGACAGCTTCGACAGGATGGAAAAGACAGTACTGAAGGTGGCGCTGGTGGGGCGCGAGGGAGCCTCCCTGCCGGTGTACTTCCTGTCCTGGCTGCAGTCCAAGCTGCTGTTCTACAAGCTGTCGGAGCTTCCCGAGGAGGAGATGGAGGACAAGCCCGTGGACTACACCAAGCTGGACAACTTCGAGATCATGCAGCGGGCCAGATGGCAGCTGGAGCGCGGCGCGGTGGAGCGGGCGGCGCGGCTGGTGGGCGCGCTGCGGGGCGCGGGGCGGGCGGCGGGCGCGGCGTGGGCGGGGCCGGCGCGCGAGCACCTGGCCGTGCGTCAGGCCGCGCACGCGCTCATGGCGCACGCCGAGCTGTCCTCGCTGCTGCACGTGTAG
Protein
MYKFTNHLSNAKLVLVRRHIDGSSAALIVLRQNSYSQDHGTRQQPRKSRKLFWSTVGATVLTGAAVVYAKHSPEARNWLESNVPWANDLVALVYQEKSSPWKFTVDRFNQATTSISHFVFGKEGVTPLEIKSRSQQDIDADAAKKATSKDFQLPPPSLEPLYVEEKKLGTPDVDTPATVVQTERCEPARAPAAAVPAELVQLERDMMDNTKLALDSYKRASQHCKRYNDALYKIVESTVEELDKRHFSALQGAQSERDAAHEQAAEAAARAKSAIQRVESALEAGAPVPPPAAAATRRHAAAFGEQLAAAEAEYCRCRDEALLADRYWDKVEAARSAFRAELAALFPGADLSARSLPSAHVDLLLVYTLKQIQFLQNQLAELQTVREQKINRAIECHDEKALIEAKVEDLIKAERVEREKEFLKRSLALQAEAHRSLREQLKKQFEIQQEVLQEKVASKEKEVMSRLARAQSERLEHERAQHKRELGAAAARLAAVQHALKQRAGAEEEARRSAALWAAAGGVLAALSAPRRAPLRDHVAAVRSAGKDDKLVQSILQSISRDALEHGVATEQELRDSFDRMEKTVLKVALVGREGASLPVYFLSWLQSKLLFYKLSELPEEEMEDKPVDYTKLDNFEIMQRARWQLERGAVERAARLVGALRGAGRAAGAAWAGPAREHLAVRQAAHALMAHAELSSLLHV
Summary
Description
Component of the MICOS complex, a large protein complex of the mitochondrial inner membrane that plays crucial roles in the maintenance of crista junctions, inner membrane architecture, and formation of contact sites to the outer membrane.
Subunit
Component of the mitochondrial contact site and cristae organizing system (MICOS) complex.
Similarity
Belongs to the MICOS complex subunit Mic60 family.
Uniprot
EMBL
AGBW02013848
OWR42660.1
RSAL01000019
RVE52792.1
KZ150122
PZC73165.1
+ More
GDQN01004832 JAT86222.1 KQ459144 KPJ03614.1 DS232669 EDS44578.1 KQ976598 KYM79575.1 ADTU01001014 ADTU01001015 ADTU01001016 KQ977800 KYM99578.1 GDKW01000212 JAI56383.1 KQ979039 KYN23262.1 KQ983002 KYQ48413.1 KQ981855 KYN34893.1
GDQN01004832 JAT86222.1 KQ459144 KPJ03614.1 DS232669 EDS44578.1 KQ976598 KYM79575.1 ADTU01001014 ADTU01001015 ADTU01001016 KQ977800 KYM99578.1 GDKW01000212 JAI56383.1 KQ979039 KYN23262.1 KQ983002 KYQ48413.1 KQ981855 KYN34893.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
Ontologies
PANTHER
Topology
Subcellular location
Mitochondrion inner membrane
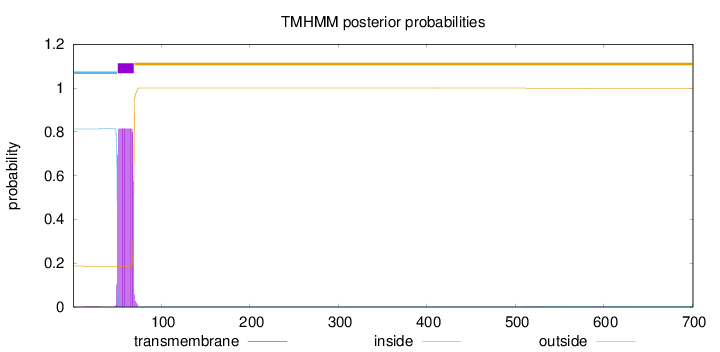
Length:
701
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
15.38533
Exp number, first 60 AAs:
8.16454
Total prob of N-in:
0.81311
inside
1 - 50
TMhelix
51 - 69
outside
70 - 701
Population Genetic Test Statistics
Pi
197.186506
Theta
161.94339
Tajima's D
0.654689
CLR
122.123812
CSRT
0.56237188140593
Interpretation
Uncertain
