Pre Gene Modal
BGIBMGA000002
Annotation
PREDICTED:_uncharacterized_protein_LOC101745378_[Bombyx_mori]
Full name
MICOS complex subunit MIC60
Alternative Name
Mitofilin
Location in the cell
Cytoplasmic Reliability : 1.605 Nuclear Reliability : 1.056
Sequence
CDS
ATGGACAGGGAGTATAGGAAACGGTCAGAGGAGCTTCGTGCTGCCAACGAGCACATGATATCGGAGTGCTTGAAGAGGCAGCGCGCGGAGCACGAGGAGACGCTTCGACAGCGCGTGCTGCAGAAGGAGATCGAGGCGAAGGCGAGATTGAACAAGTTGATTGCCGAGAAGGTCGCGGCCGAGAAGCTGGTGCTCGCGGCGCAGCTCGCGGAGATGAGGAGCAAACTGAACGTGGTCGAGGAGAAACTCAACGGTATAGATTTTTTTTTATTATTTAGACGGGTGGACGAACTTACAGCCCACCTGGTGTTAAGCTGTTACTGGAGCCCATAG
Protein
MDREYRKRSEELRAANEHMISECLKRQRAEHEETLRQRVLQKEIEAKARLNKLIAEKVAAEKLVLAAQLAEMRSKLNVVEEKLNGIDFFLLFRRVDELTAHLVLSCYWSP
Summary
Description
Component of the MICOS complex, a large protein complex of the mitochondrial inner membrane that plays crucial roles in the maintenance of crista junctions, inner membrane architecture, and formation of contact sites to the outer membrane.
Subunit
Component of the mitochondrial contact site and cristae organizing system (MICOS) complex.
Similarity
Belongs to the MICOS complex subunit Mic60 family.
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF09731 Mitofilin
Interpro
IPR019133
Mt-IM_prot_Mitofilin
ProteinModelPortal
Ontologies
PANTHER
Topology
Subcellular location
Mitochondrion inner membrane
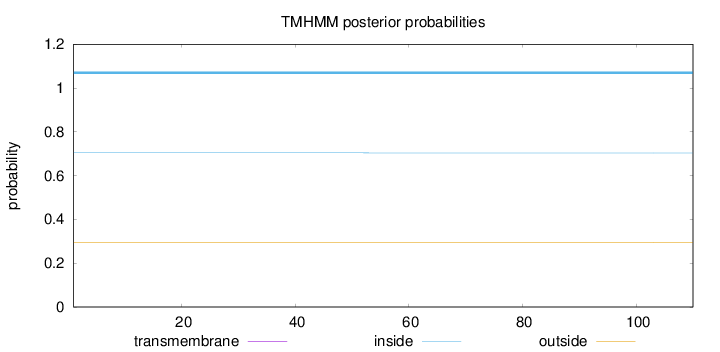
Length:
110
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00199
Exp number, first 60 AAs:
0.00028
Total prob of N-in:
0.70393
inside
1 - 110
Population Genetic Test Statistics
Pi
228.332357
Theta
162.773212
Tajima's D
1.198475
CLR
0.000285
CSRT
0.709614519274036
Interpretation
Uncertain
