Gene
KWMTBOMO15843
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Full name
Craniofacial development protein 2
Alternative Name
p97 bucentaur protein
Location in the cell
Nuclear Reliability : 2.075
Sequence
CDS
ATGATCAATGATGACCGTATTCGCAACATTGTGGGAAGGTTTGGTGTGGGTTCTAAGAGCAATCGTGGCCAACGACTTGTGGAGTATCGCATCGAGAGGAATCTTTGCATAACAAACACGCAATTCCAACATCACCCAAGACGTTTATACACATGGACTTTCCCTGGAGCGCGGTATAGCAATCAGATCGATCACATACTCGTGAACACGCGTTGGAAGTCGTCCGTCGTAAATGTGAAGACATGCTTTTTTAATTTGCGCTCAGGCTAA
Protein
MINDDRIRNIVGRFGVGSKSNRGQRLVEYRIERNLCITNTQFQHHPRRLYTWTFPGARYSNQIDHILVNTRWKSSVVNVKTCFFNLRSG
Summary
Miscellaneous
Gene duplication of the ancestral BCNT gene leads to the h-type BCNT (CFDP1) gene and the p97BCNT (CFDP2) gene. The latter contains a region derived from the endonuclease domain of a retrotransposable element RTE-1. This repetitive sequence associated with the BCNT gene is specific to Ruminantia.
Keywords
Complete proteome
Cytoplasm
Direct protein sequencing
Nucleus
Reference proteome
Feature
chain Craniofacial development protein 2
Uniprot
A0A2H1W4B8
D7F163
D7F174
D7F168
D7F162
A0A2H1V4P4
+ More
D7F172 A0A2W1BNH4 A0A3D1L4K7 A0A2W1BHL0 A0A2S2NUN8 A0A0L8I0S1 A0A0L8GIP3 A0A023GCB5 D7F175 A0A0J7L4A4 A0A023GD30 E9I958 W4XIJ4 L8IKR7 A0A3Q1M0C5 A0A0J7NGC2 L8IQH5 W4YZ24 A0JBZ9 A0A1S3DE02 L8IMU7 B6F238 F1MS40 A0A3Q1MSH0 O02751 A0A2G8JL44 A0A3Q1NFT8 L8IKL3 A0A0B1PP52 A0A3Q1MMW9 A0A3Q1NNM3 W4Y7C9 A0A3Q1M3D8 A0A0B7BS70 Q4ADK3 A0A0B7BR26 A0A0B7BTN4 H2YWZ5 A0A3Q1MMZ0 A0A3Q1MLE5 A0A3Q1MHP3 A0A0L8HFQ4 W4Z0F5 D7F161 A0A3Q1LYK9 A0A3Q1NNC3 A0A0B7BTM9 A0A0B7BU90 A0A3Q1MD43 A0A0B7BTM4 A0A0B7BR31 A0A0B7BUA0 W4Z830 A0A0B7BR36 A0A0B7BRV7 A0A3Q1EK63 A0A0B7BRW7 W4YPR9 A0A3Q1MC12 A0A3Q1MD68 A0A3Q1MTP7 A0A3Q1LUJ6 Q8SQ39 Q7YRU5 D5LB39 A0A3Q1MEL3 A0A2G8KUE0 A0A3Q1LQW1 Q588U8 A0A1S3DQR0 A0A3Q1M1R5 A0A0B7B4J6 A0A3Q1MS18 A0A3Q1MM58 A0A3Q1MVK1 A0A3Q1M6I3 A0A3Q1MM21 A0A0B7B1J3 A0A0B7BTK3 L8I6X2 D7F167 A0A3Q1M0B3 A0A3Q1MQA7 A0A3S3P648 A0A0P4VZ55 W4Y522 A0A0B7B2E7 A0A3Q1M255 A0A1S3DNU9 A0A3Q1LMM1 Q4R2S8 A0A3Q1NGA3 W4XDX3 J9KN98
D7F172 A0A2W1BNH4 A0A3D1L4K7 A0A2W1BHL0 A0A2S2NUN8 A0A0L8I0S1 A0A0L8GIP3 A0A023GCB5 D7F175 A0A0J7L4A4 A0A023GD30 E9I958 W4XIJ4 L8IKR7 A0A3Q1M0C5 A0A0J7NGC2 L8IQH5 W4YZ24 A0JBZ9 A0A1S3DE02 L8IMU7 B6F238 F1MS40 A0A3Q1MSH0 O02751 A0A2G8JL44 A0A3Q1NFT8 L8IKL3 A0A0B1PP52 A0A3Q1MMW9 A0A3Q1NNM3 W4Y7C9 A0A3Q1M3D8 A0A0B7BS70 Q4ADK3 A0A0B7BR26 A0A0B7BTN4 H2YWZ5 A0A3Q1MMZ0 A0A3Q1MLE5 A0A3Q1MHP3 A0A0L8HFQ4 W4Z0F5 D7F161 A0A3Q1LYK9 A0A3Q1NNC3 A0A0B7BTM9 A0A0B7BU90 A0A3Q1MD43 A0A0B7BTM4 A0A0B7BR31 A0A0B7BUA0 W4Z830 A0A0B7BR36 A0A0B7BRV7 A0A3Q1EK63 A0A0B7BRW7 W4YPR9 A0A3Q1MC12 A0A3Q1MD68 A0A3Q1MTP7 A0A3Q1LUJ6 Q8SQ39 Q7YRU5 D5LB39 A0A3Q1MEL3 A0A2G8KUE0 A0A3Q1LQW1 Q588U8 A0A1S3DQR0 A0A3Q1M1R5 A0A0B7B4J6 A0A3Q1MS18 A0A3Q1MM58 A0A3Q1MVK1 A0A3Q1M6I3 A0A3Q1MM21 A0A0B7B1J3 A0A0B7BTK3 L8I6X2 D7F167 A0A3Q1M0B3 A0A3Q1MQA7 A0A3S3P648 A0A0P4VZ55 W4Y522 A0A0B7B2E7 A0A3Q1M255 A0A1S3DNU9 A0A3Q1LMM1 Q4R2S8 A0A3Q1NGA3 W4XDX3 J9KN98
Pubmed
EMBL
ODYU01006118
SOQ47682.1
FJ265548
ADI61816.1
FJ265559
ADI61827.1
+ More
FJ265553 ADI61821.1 FJ265547 ADI61815.1 ODYU01000634 SOQ35746.1 FJ265557 ADI61825.1 KZ150025 PZC74827.1 DPCO01000033 HCE59366.1 KZ150087 PZC73751.1 GGMR01007827 MBY20446.1 KQ416818 KOF95029.1 KQ421665 KOF76833.1 GBBM01003916 JAC31502.1 FJ265560 ADI61828.1 LBMM01000801 KMQ97461.1 GBBM01003639 JAC31779.1 GL761727 EFZ22895.1 AAGJ04122993 JH881209 ELR55722.1 LBMM01005351 KMQ91585.1 JH880989 ELR57387.1 AAGJ04003947 BC149202 AB259013 AAI49203.1 BAF36506.1 ELR57388.1 LWLT01000020 AB465690 BAG75458.1 D84513 D84514 D84515 AB081095 AB005652 MRZV01001667 PIK36482.1 JH881217 ELR55677.1 KN538403 KHJ42451.1 AAGJ04113592 AAGJ04113593 AAGJ04113594 HACG01048887 CEK95752.1 AB213486 BAE19808.1 HACG01048869 CEK95734.1 HACG01048886 CEK95751.1 KQ418263 KOF88093.1 AAGJ04089624 FJ265546 ADI61814.1 HACG01048881 CEK95746.1 HACG01048875 CEK95740.1 HACG01048876 CEK95741.1 HACG01048874 CEK95739.1 HACG01048885 CEK95750.1 AAGJ04154883 HACG01048879 CEK95744.1 HACG01048873 CEK95738.1 HACG01048883 CEK95748.1 AAGJ04030456 AB084085 BAB90842.1 AB113247 BAC78521.1 GU815090 ADF18553.1 MRZV01000365 PIK51575.1 AB191483 AB192410 AB158226 HACG01040351 CEK87216.1 HACG01040349 CEK87214.1 HACG01048595 CEK95460.1 JH881842 ELR51968.1 FJ265552 ADI61820.1 NCKU01004395 RWS06003.1 GDRN01105844 GDRN01105842 JAI57667.1 AAGJ04102956 HACG01040353 CEK87218.1 AB190444 BAE00011.1 ABLF02014119 ABLF02061067
FJ265553 ADI61821.1 FJ265547 ADI61815.1 ODYU01000634 SOQ35746.1 FJ265557 ADI61825.1 KZ150025 PZC74827.1 DPCO01000033 HCE59366.1 KZ150087 PZC73751.1 GGMR01007827 MBY20446.1 KQ416818 KOF95029.1 KQ421665 KOF76833.1 GBBM01003916 JAC31502.1 FJ265560 ADI61828.1 LBMM01000801 KMQ97461.1 GBBM01003639 JAC31779.1 GL761727 EFZ22895.1 AAGJ04122993 JH881209 ELR55722.1 LBMM01005351 KMQ91585.1 JH880989 ELR57387.1 AAGJ04003947 BC149202 AB259013 AAI49203.1 BAF36506.1 ELR57388.1 LWLT01000020 AB465690 BAG75458.1 D84513 D84514 D84515 AB081095 AB005652 MRZV01001667 PIK36482.1 JH881217 ELR55677.1 KN538403 KHJ42451.1 AAGJ04113592 AAGJ04113593 AAGJ04113594 HACG01048887 CEK95752.1 AB213486 BAE19808.1 HACG01048869 CEK95734.1 HACG01048886 CEK95751.1 KQ418263 KOF88093.1 AAGJ04089624 FJ265546 ADI61814.1 HACG01048881 CEK95746.1 HACG01048875 CEK95740.1 HACG01048876 CEK95741.1 HACG01048874 CEK95739.1 HACG01048885 CEK95750.1 AAGJ04154883 HACG01048879 CEK95744.1 HACG01048873 CEK95738.1 HACG01048883 CEK95748.1 AAGJ04030456 AB084085 BAB90842.1 AB113247 BAC78521.1 GU815090 ADF18553.1 MRZV01000365 PIK51575.1 AB191483 AB192410 AB158226 HACG01040351 CEK87216.1 HACG01040349 CEK87214.1 HACG01048595 CEK95460.1 JH881842 ELR51968.1 FJ265552 ADI61820.1 NCKU01004395 RWS06003.1 GDRN01105844 GDRN01105842 JAI57667.1 AAGJ04102956 HACG01040353 CEK87218.1 AB190444 BAE00011.1 ABLF02014119 ABLF02061067
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
A0A2H1W4B8
D7F163
D7F174
D7F168
D7F162
A0A2H1V4P4
+ More
D7F172 A0A2W1BNH4 A0A3D1L4K7 A0A2W1BHL0 A0A2S2NUN8 A0A0L8I0S1 A0A0L8GIP3 A0A023GCB5 D7F175 A0A0J7L4A4 A0A023GD30 E9I958 W4XIJ4 L8IKR7 A0A3Q1M0C5 A0A0J7NGC2 L8IQH5 W4YZ24 A0JBZ9 A0A1S3DE02 L8IMU7 B6F238 F1MS40 A0A3Q1MSH0 O02751 A0A2G8JL44 A0A3Q1NFT8 L8IKL3 A0A0B1PP52 A0A3Q1MMW9 A0A3Q1NNM3 W4Y7C9 A0A3Q1M3D8 A0A0B7BS70 Q4ADK3 A0A0B7BR26 A0A0B7BTN4 H2YWZ5 A0A3Q1MMZ0 A0A3Q1MLE5 A0A3Q1MHP3 A0A0L8HFQ4 W4Z0F5 D7F161 A0A3Q1LYK9 A0A3Q1NNC3 A0A0B7BTM9 A0A0B7BU90 A0A3Q1MD43 A0A0B7BTM4 A0A0B7BR31 A0A0B7BUA0 W4Z830 A0A0B7BR36 A0A0B7BRV7 A0A3Q1EK63 A0A0B7BRW7 W4YPR9 A0A3Q1MC12 A0A3Q1MD68 A0A3Q1MTP7 A0A3Q1LUJ6 Q8SQ39 Q7YRU5 D5LB39 A0A3Q1MEL3 A0A2G8KUE0 A0A3Q1LQW1 Q588U8 A0A1S3DQR0 A0A3Q1M1R5 A0A0B7B4J6 A0A3Q1MS18 A0A3Q1MM58 A0A3Q1MVK1 A0A3Q1M6I3 A0A3Q1MM21 A0A0B7B1J3 A0A0B7BTK3 L8I6X2 D7F167 A0A3Q1M0B3 A0A3Q1MQA7 A0A3S3P648 A0A0P4VZ55 W4Y522 A0A0B7B2E7 A0A3Q1M255 A0A1S3DNU9 A0A3Q1LMM1 Q4R2S8 A0A3Q1NGA3 W4XDX3 J9KN98
D7F172 A0A2W1BNH4 A0A3D1L4K7 A0A2W1BHL0 A0A2S2NUN8 A0A0L8I0S1 A0A0L8GIP3 A0A023GCB5 D7F175 A0A0J7L4A4 A0A023GD30 E9I958 W4XIJ4 L8IKR7 A0A3Q1M0C5 A0A0J7NGC2 L8IQH5 W4YZ24 A0JBZ9 A0A1S3DE02 L8IMU7 B6F238 F1MS40 A0A3Q1MSH0 O02751 A0A2G8JL44 A0A3Q1NFT8 L8IKL3 A0A0B1PP52 A0A3Q1MMW9 A0A3Q1NNM3 W4Y7C9 A0A3Q1M3D8 A0A0B7BS70 Q4ADK3 A0A0B7BR26 A0A0B7BTN4 H2YWZ5 A0A3Q1MMZ0 A0A3Q1MLE5 A0A3Q1MHP3 A0A0L8HFQ4 W4Z0F5 D7F161 A0A3Q1LYK9 A0A3Q1NNC3 A0A0B7BTM9 A0A0B7BU90 A0A3Q1MD43 A0A0B7BTM4 A0A0B7BR31 A0A0B7BUA0 W4Z830 A0A0B7BR36 A0A0B7BRV7 A0A3Q1EK63 A0A0B7BRW7 W4YPR9 A0A3Q1MC12 A0A3Q1MD68 A0A3Q1MTP7 A0A3Q1LUJ6 Q8SQ39 Q7YRU5 D5LB39 A0A3Q1MEL3 A0A2G8KUE0 A0A3Q1LQW1 Q588U8 A0A1S3DQR0 A0A3Q1M1R5 A0A0B7B4J6 A0A3Q1MS18 A0A3Q1MM58 A0A3Q1MVK1 A0A3Q1M6I3 A0A3Q1MM21 A0A0B7B1J3 A0A0B7BTK3 L8I6X2 D7F167 A0A3Q1M0B3 A0A3Q1MQA7 A0A3S3P648 A0A0P4VZ55 W4Y522 A0A0B7B2E7 A0A3Q1M255 A0A1S3DNU9 A0A3Q1LMM1 Q4R2S8 A0A3Q1NGA3 W4XDX3 J9KN98
Ontologies
PANTHER
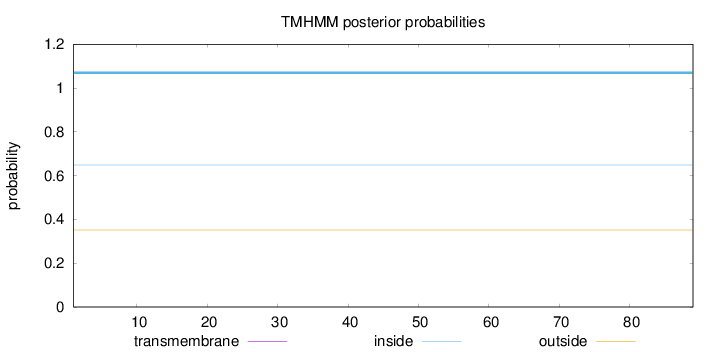
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
Length:
89
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00059
Exp number, first 60 AAs:
0.00024
Total prob of N-in:
0.64890
inside
1 - 89
Population Genetic Test Statistics
Pi
198.483031
Theta
143.839923
Tajima's D
1.193927
CLR
0
CSRT
0.702614869256537
Interpretation
Uncertain
