Gene
KWMTBOMO15839
Pre Gene Modal
BGIBMGA000057
Annotation
hypothetical_protein_KGM_14251_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 1.879
Sequence
CDS
ATGGCGAACAACAGAATCCCTTCAGGACCTAACACTCCTGGCAGTCAGCAGACTCCCACACAGCAGGGAATCAAAATATTCATACAAAGAGACTATTCTGAAGGCACTGCAGTTAAATTCCAGACTAGATTCCCGCCTGAGCTTGAAGAAAGGATAGACAGACAAACCTTTGAACATACAATAGAGAGACTGAATGAGCACTTTGAAATGGCAGAGACAGCAGACTGCAGTACATATTGTGAGGGATGCCTTGCCTGTCTGACTGCCTACTTCATTTATATCTGTACAGAGACACATTATGAAAAGCATTTAAGGAAAGTCTCAAAATTCATAGCAACACAAAACGAAAGGGTATACAACCCCCGAGGTGTTCATATAACTGATCCCATTCTTCGCGGACTACGAGTCATTGAAATCACGATGATAGACGTACCCAACTGCCAGAGCCCGACAGGGAATAACACATCGATCCGACATACTTGA
Protein
MANNRIPSGPNTPGSQQTPTQQGIKIFIQRDYSEGTAVKFQTRFPPELEERIDRQTFEHTIERLNEHFEMAETADCSTYCEGCLACLTAYFIYICTETHYEKHLRKVSKFIATQNERVYNPRGVHITDPILRGLRVIEITMIDVPNCQSPTGNNTSIRHT
Summary
Uniprot
H9IS31
A0A2A4IW21
A0A2H1V5W7
A0A2W1C067
A0A212F9I7
A0A0N0PER1
+ More
A0A194QF99 A0A1E1WRG0 Q16IA1 A0A023EGL8 A0A1Q3F6R4 B0WJ66 A0A182TMB2 A0A182QIX7 A0A182YGH9 A0A182M631 A0A182P5Z1 A0A182KDK1 A0A182XIY2 Q7PQN9 A0A182I7X5 A0A084W397 A0A182J256 A0A2M4AWR6 A0A2M4C2K9 W5JQU5 A0A182F6N9 A0A182N5N1 U5ET72 A0A1W4X912 A0A0T6AZI1 A0A182W0J1 A0A1L8DW79 A0A1L8DW50 A0A1Y1LM13 V9IBP3 A0A088ADE0 A0A1L8DW65 A0A1L8DW76 A0A2A3EDJ6 V5H1T7 A0A336M8E5 A0A0N0U4Y1 A0A182RYI5 A0A0K8TLR5 D6X0V0 A0A0L7RJW1 A0A232FHH8 K7IM96 A0A1B0GK53 A0A067R3X3 A0A034WWS5 A0A0K8V688 W8CCQ9 A0A0A1XQ91 A0A034WVL7 A0A310SF30 A0A1A9YQ45 A0A1B0B4W3 A0A1A9ZSZ6 A0A1I8N1H2 D3TPA9 A0A1A9UCX5 B4LVU3 A0A0L0C9R2 A0A1A9W5L4 A0A1I8PT57 A0A0P4VPP3 A0A0V0GCY6 A0A069DP35 B4K9M1 A0A154PS93 E2AIP8 E2BNJ9 E9IWT4 A0A0J7L3D7 A0A026X3F4 A0A0A9Y7T3 B4NAL3 A0A1B6M5Y5 A0A0K8SC18 A0A3L8DKW9 A0A151IWD8 A0A151WMX8 A0A1B6CDJ1 B3M0I9 A0A1B6GNJ6 A0A1B6HNV8 F4WLE5 A0A0M4ES83 A0A195D5K2 A0A224XMW1 B4JXW9 A0A1W4VLP7 B4ICL9 B3P675 Q9VBF0 B4PSG5 A0A0B4KH14
A0A194QF99 A0A1E1WRG0 Q16IA1 A0A023EGL8 A0A1Q3F6R4 B0WJ66 A0A182TMB2 A0A182QIX7 A0A182YGH9 A0A182M631 A0A182P5Z1 A0A182KDK1 A0A182XIY2 Q7PQN9 A0A182I7X5 A0A084W397 A0A182J256 A0A2M4AWR6 A0A2M4C2K9 W5JQU5 A0A182F6N9 A0A182N5N1 U5ET72 A0A1W4X912 A0A0T6AZI1 A0A182W0J1 A0A1L8DW79 A0A1L8DW50 A0A1Y1LM13 V9IBP3 A0A088ADE0 A0A1L8DW65 A0A1L8DW76 A0A2A3EDJ6 V5H1T7 A0A336M8E5 A0A0N0U4Y1 A0A182RYI5 A0A0K8TLR5 D6X0V0 A0A0L7RJW1 A0A232FHH8 K7IM96 A0A1B0GK53 A0A067R3X3 A0A034WWS5 A0A0K8V688 W8CCQ9 A0A0A1XQ91 A0A034WVL7 A0A310SF30 A0A1A9YQ45 A0A1B0B4W3 A0A1A9ZSZ6 A0A1I8N1H2 D3TPA9 A0A1A9UCX5 B4LVU3 A0A0L0C9R2 A0A1A9W5L4 A0A1I8PT57 A0A0P4VPP3 A0A0V0GCY6 A0A069DP35 B4K9M1 A0A154PS93 E2AIP8 E2BNJ9 E9IWT4 A0A0J7L3D7 A0A026X3F4 A0A0A9Y7T3 B4NAL3 A0A1B6M5Y5 A0A0K8SC18 A0A3L8DKW9 A0A151IWD8 A0A151WMX8 A0A1B6CDJ1 B3M0I9 A0A1B6GNJ6 A0A1B6HNV8 F4WLE5 A0A0M4ES83 A0A195D5K2 A0A224XMW1 B4JXW9 A0A1W4VLP7 B4ICL9 B3P675 Q9VBF0 B4PSG5 A0A0B4KH14
Pubmed
19121390
28756777
22118469
26354079
17510324
24945155
+ More
26483478 25244985 12364791 14747013 17210077 24438588 20920257 23761445 28004739 26369729 18362917 19820115 28648823 20075255 24845553 25348373 24495485 25830018 25315136 20353571 17994087 26108605 27129103 26334808 20798317 21282665 24508170 25401762 26823975 30249741 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
26483478 25244985 12364791 14747013 17210077 24438588 20920257 23761445 28004739 26369729 18362917 19820115 28648823 20075255 24845553 25348373 24495485 25830018 25315136 20353571 17994087 26108605 27129103 26334808 20798317 21282665 24508170 25401762 26823975 30249741 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
EMBL
BABH01035664
BABH01035665
NWSH01005798
PCG63941.1
ODYU01000846
SOQ36221.1
+ More
KZ149905 PZC78366.1 AGBW02009573 OWR50402.1 KQ459717 KPJ20256.1 KQ459144 KPJ03620.1 GDQN01001439 JAT89615.1 CH478094 EAT33989.1 JXUM01139118 GAPW01005647 KQ568846 JAC07951.1 KXJ68862.1 GFDL01011795 JAV23250.1 DS231956 EDS28956.1 AXCN02001159 AXCM01008033 AAAB01008880 EAA08561.5 APCN01002573 ATLV01019871 KE525281 KFB44691.1 AXCP01003717 GGFK01011899 MBW45220.1 GGFJ01010392 MBW59533.1 ADMH02000440 ETN66476.1 GANO01002910 JAB56961.1 LJIG01022456 KRT80497.1 GFDF01003396 JAV10688.1 GFDF01003414 JAV10670.1 GEZM01051824 GEZM01051823 JAV74682.1 JR039081 AEY58520.1 GFDF01003397 JAV10687.1 GFDF01003398 JAV10686.1 KZ288271 PBC29815.1 GALX01000239 JAB68227.1 UFQS01000690 UFQT01000690 SSX06238.1 SSX26592.1 KQ435812 KOX72732.1 GDAI01002296 JAI15307.1 KQ971372 EFA10556.1 KQ414581 KOC71026.1 NNAY01000222 OXU29909.1 AJWK01022591 AJWK01022592 KK852720 KDR17757.1 GAKP01000282 JAC58670.1 GDHF01017867 JAI34447.1 GAMC01003266 JAC03290.1 GBXI01000798 JAD13494.1 GAKP01000283 JAC58669.1 KQ760549 OAD59891.1 JXJN01008560 CCAG010011760 EZ423261 ADD19537.1 CH940650 EDW67548.2 JRES01000718 KNC28991.1 GDKW01003452 JAI53143.1 GECL01000355 JAP05769.1 GBGD01003463 JAC85426.1 CH933806 EDW16681.2 KQ435092 KZC14617.1 GL439853 EFN66685.1 GL449414 EFN82778.1 GL766597 EFZ15010.1 LBMM01000974 KMQ97128.1 KK107019 EZA62797.1 GBHO01016456 GDHC01015759 JAG27148.1 JAQ02870.1 CH964232 EDW80827.2 GEBQ01009853 GEBQ01008645 JAT30124.1 JAT31332.1 GBRD01014980 JAG50846.1 QOIP01000007 RLU20956.1 KQ980859 KYN12135.1 KQ982944 KYQ49045.1 GEDC01025820 GEDC01006493 JAS11478.1 JAS30805.1 CH902617 EDV44236.2 GECZ01005846 JAS63923.1 GECU01031411 JAS76295.1 GL888207 EGI65108.1 CP012526 ALC45430.1 KQ976870 KYN07719.1 GFTR01002589 JAW13837.1 CH916377 EDV90531.1 CH480828 EDW45115.1 CH954182 EDV53545.1 AE014297 AY061563 AY071519 AAF56592.1 AAL29111.1 AAL49141.1 CM000160 EDW98627.2 AGB96366.1
KZ149905 PZC78366.1 AGBW02009573 OWR50402.1 KQ459717 KPJ20256.1 KQ459144 KPJ03620.1 GDQN01001439 JAT89615.1 CH478094 EAT33989.1 JXUM01139118 GAPW01005647 KQ568846 JAC07951.1 KXJ68862.1 GFDL01011795 JAV23250.1 DS231956 EDS28956.1 AXCN02001159 AXCM01008033 AAAB01008880 EAA08561.5 APCN01002573 ATLV01019871 KE525281 KFB44691.1 AXCP01003717 GGFK01011899 MBW45220.1 GGFJ01010392 MBW59533.1 ADMH02000440 ETN66476.1 GANO01002910 JAB56961.1 LJIG01022456 KRT80497.1 GFDF01003396 JAV10688.1 GFDF01003414 JAV10670.1 GEZM01051824 GEZM01051823 JAV74682.1 JR039081 AEY58520.1 GFDF01003397 JAV10687.1 GFDF01003398 JAV10686.1 KZ288271 PBC29815.1 GALX01000239 JAB68227.1 UFQS01000690 UFQT01000690 SSX06238.1 SSX26592.1 KQ435812 KOX72732.1 GDAI01002296 JAI15307.1 KQ971372 EFA10556.1 KQ414581 KOC71026.1 NNAY01000222 OXU29909.1 AJWK01022591 AJWK01022592 KK852720 KDR17757.1 GAKP01000282 JAC58670.1 GDHF01017867 JAI34447.1 GAMC01003266 JAC03290.1 GBXI01000798 JAD13494.1 GAKP01000283 JAC58669.1 KQ760549 OAD59891.1 JXJN01008560 CCAG010011760 EZ423261 ADD19537.1 CH940650 EDW67548.2 JRES01000718 KNC28991.1 GDKW01003452 JAI53143.1 GECL01000355 JAP05769.1 GBGD01003463 JAC85426.1 CH933806 EDW16681.2 KQ435092 KZC14617.1 GL439853 EFN66685.1 GL449414 EFN82778.1 GL766597 EFZ15010.1 LBMM01000974 KMQ97128.1 KK107019 EZA62797.1 GBHO01016456 GDHC01015759 JAG27148.1 JAQ02870.1 CH964232 EDW80827.2 GEBQ01009853 GEBQ01008645 JAT30124.1 JAT31332.1 GBRD01014980 JAG50846.1 QOIP01000007 RLU20956.1 KQ980859 KYN12135.1 KQ982944 KYQ49045.1 GEDC01025820 GEDC01006493 JAS11478.1 JAS30805.1 CH902617 EDV44236.2 GECZ01005846 JAS63923.1 GECU01031411 JAS76295.1 GL888207 EGI65108.1 CP012526 ALC45430.1 KQ976870 KYN07719.1 GFTR01002589 JAW13837.1 CH916377 EDV90531.1 CH480828 EDW45115.1 CH954182 EDV53545.1 AE014297 AY061563 AY071519 AAF56592.1 AAL29111.1 AAL49141.1 CM000160 EDW98627.2 AGB96366.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000008820
+ More
UP000069940 UP000249989 UP000002320 UP000075902 UP000075886 UP000076408 UP000075883 UP000075885 UP000075881 UP000076407 UP000007062 UP000075840 UP000030765 UP000075880 UP000000673 UP000069272 UP000075884 UP000192223 UP000075920 UP000005203 UP000242457 UP000053105 UP000075900 UP000007266 UP000053825 UP000215335 UP000002358 UP000092461 UP000027135 UP000092443 UP000092460 UP000092445 UP000095301 UP000092444 UP000078200 UP000008792 UP000037069 UP000091820 UP000095300 UP000009192 UP000076502 UP000000311 UP000008237 UP000036403 UP000053097 UP000007798 UP000279307 UP000078492 UP000075809 UP000007801 UP000007755 UP000092553 UP000078542 UP000001070 UP000192221 UP000001292 UP000008711 UP000000803 UP000002282
UP000069940 UP000249989 UP000002320 UP000075902 UP000075886 UP000076408 UP000075883 UP000075885 UP000075881 UP000076407 UP000007062 UP000075840 UP000030765 UP000075880 UP000000673 UP000069272 UP000075884 UP000192223 UP000075920 UP000005203 UP000242457 UP000053105 UP000075900 UP000007266 UP000053825 UP000215335 UP000002358 UP000092461 UP000027135 UP000092443 UP000092460 UP000092445 UP000095301 UP000092444 UP000078200 UP000008792 UP000037069 UP000091820 UP000095300 UP000009192 UP000076502 UP000000311 UP000008237 UP000036403 UP000053097 UP000007798 UP000279307 UP000078492 UP000075809 UP000007801 UP000007755 UP000092553 UP000078542 UP000001070 UP000192221 UP000001292 UP000008711 UP000000803 UP000002282
Pfam
PF10256 Erf4
Interpro
IPR019383
Golgin_A_7/ERF4
ProteinModelPortal
H9IS31
A0A2A4IW21
A0A2H1V5W7
A0A2W1C067
A0A212F9I7
A0A0N0PER1
+ More
A0A194QF99 A0A1E1WRG0 Q16IA1 A0A023EGL8 A0A1Q3F6R4 B0WJ66 A0A182TMB2 A0A182QIX7 A0A182YGH9 A0A182M631 A0A182P5Z1 A0A182KDK1 A0A182XIY2 Q7PQN9 A0A182I7X5 A0A084W397 A0A182J256 A0A2M4AWR6 A0A2M4C2K9 W5JQU5 A0A182F6N9 A0A182N5N1 U5ET72 A0A1W4X912 A0A0T6AZI1 A0A182W0J1 A0A1L8DW79 A0A1L8DW50 A0A1Y1LM13 V9IBP3 A0A088ADE0 A0A1L8DW65 A0A1L8DW76 A0A2A3EDJ6 V5H1T7 A0A336M8E5 A0A0N0U4Y1 A0A182RYI5 A0A0K8TLR5 D6X0V0 A0A0L7RJW1 A0A232FHH8 K7IM96 A0A1B0GK53 A0A067R3X3 A0A034WWS5 A0A0K8V688 W8CCQ9 A0A0A1XQ91 A0A034WVL7 A0A310SF30 A0A1A9YQ45 A0A1B0B4W3 A0A1A9ZSZ6 A0A1I8N1H2 D3TPA9 A0A1A9UCX5 B4LVU3 A0A0L0C9R2 A0A1A9W5L4 A0A1I8PT57 A0A0P4VPP3 A0A0V0GCY6 A0A069DP35 B4K9M1 A0A154PS93 E2AIP8 E2BNJ9 E9IWT4 A0A0J7L3D7 A0A026X3F4 A0A0A9Y7T3 B4NAL3 A0A1B6M5Y5 A0A0K8SC18 A0A3L8DKW9 A0A151IWD8 A0A151WMX8 A0A1B6CDJ1 B3M0I9 A0A1B6GNJ6 A0A1B6HNV8 F4WLE5 A0A0M4ES83 A0A195D5K2 A0A224XMW1 B4JXW9 A0A1W4VLP7 B4ICL9 B3P675 Q9VBF0 B4PSG5 A0A0B4KH14
A0A194QF99 A0A1E1WRG0 Q16IA1 A0A023EGL8 A0A1Q3F6R4 B0WJ66 A0A182TMB2 A0A182QIX7 A0A182YGH9 A0A182M631 A0A182P5Z1 A0A182KDK1 A0A182XIY2 Q7PQN9 A0A182I7X5 A0A084W397 A0A182J256 A0A2M4AWR6 A0A2M4C2K9 W5JQU5 A0A182F6N9 A0A182N5N1 U5ET72 A0A1W4X912 A0A0T6AZI1 A0A182W0J1 A0A1L8DW79 A0A1L8DW50 A0A1Y1LM13 V9IBP3 A0A088ADE0 A0A1L8DW65 A0A1L8DW76 A0A2A3EDJ6 V5H1T7 A0A336M8E5 A0A0N0U4Y1 A0A182RYI5 A0A0K8TLR5 D6X0V0 A0A0L7RJW1 A0A232FHH8 K7IM96 A0A1B0GK53 A0A067R3X3 A0A034WWS5 A0A0K8V688 W8CCQ9 A0A0A1XQ91 A0A034WVL7 A0A310SF30 A0A1A9YQ45 A0A1B0B4W3 A0A1A9ZSZ6 A0A1I8N1H2 D3TPA9 A0A1A9UCX5 B4LVU3 A0A0L0C9R2 A0A1A9W5L4 A0A1I8PT57 A0A0P4VPP3 A0A0V0GCY6 A0A069DP35 B4K9M1 A0A154PS93 E2AIP8 E2BNJ9 E9IWT4 A0A0J7L3D7 A0A026X3F4 A0A0A9Y7T3 B4NAL3 A0A1B6M5Y5 A0A0K8SC18 A0A3L8DKW9 A0A151IWD8 A0A151WMX8 A0A1B6CDJ1 B3M0I9 A0A1B6GNJ6 A0A1B6HNV8 F4WLE5 A0A0M4ES83 A0A195D5K2 A0A224XMW1 B4JXW9 A0A1W4VLP7 B4ICL9 B3P675 Q9VBF0 B4PSG5 A0A0B4KH14
Ontologies
GO
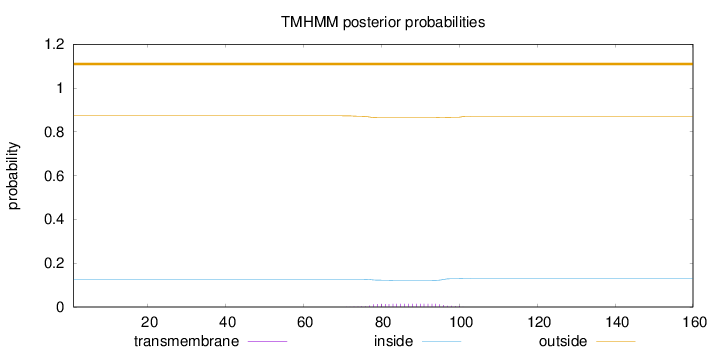
Topology
Length:
160
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.29373
Exp number, first 60 AAs:
0
Total prob of N-in:
0.12680
outside
1 - 160
Population Genetic Test Statistics
Pi
182.330127
Theta
160.613021
Tajima's D
0.39377
CLR
0
CSRT
0.484225788710564
Interpretation
Uncertain
