Gene
KWMTBOMO15834
Pre Gene Modal
BGIBMGA000006
Annotation
osiris_18_precursor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.233 Mitochondrial Reliability : 1.22
Sequence
CDS
ATGGCCAGAATAATAGTGTTGTTATCGATCGTTGCCTTCGCAAGTGCCACGTATCCGGCATCGAAACTAGTCAAGAACATTTATAACGAATGCCTTAGCCAGTATTCGGTGGAATGCGTGAAGCCGAGGACCCTGCAGTGGATGTCATCGGTTGCCAACGACGACGAAATTAAGATCACTGAAGATCTTTCTATCGTGAAGACCGGAACAGTTGAGGATGATGAATCTGCCGACCCTAGACTGGCCAAGGATCCGGCCTACGAAATGTTCGACAAAGTGGACAAGTTTCTCCAGAGTCATACTCTTAGGGTGAAAGTTCCAGAGGAGATAACGAAAAGTGCTGCTTCAGAGTACGTGCCAAGGTCTCTCTTGACTGACTTGCCTTCTGAGCTGGACATGCCCTTAGATGGAGAAGATGAGGCTGAAGTCGTTGAGGGCAGGAAGAAGAAGATCAAGCTCCCTAAACCATTGAGGATCAAGAGCAAACACGGTTTCATCAAGAAGGTTATCCTGCCTTTCCTGCTGGGTCTCAAATTTAAAGCTTCTGTTCTCGTTCCCTTGGCGCTAGCCCTCATCGCTCTGAAGACCTGGAAGGCTCTTACTCTTGGTCTCATTTCCCTTGTTTTATCTGGTGCCATGGTCATCTTCAAATTCACGAAGCCCAAGGTCGTGAACTACGAAGTAATCCACTATCCTCAACACCATGTGGAGCACCACGTAGACCACCACGCCCCTGCCTGGGACGCACACGGCCCCTACCAAGCGAGATCCTACGAAGACGCTCACGAAATCGCATACTCCGCACAAATATAA
Protein
MARIIVLLSIVAFASATYPASKLVKNIYNECLSQYSVECVKPRTLQWMSSVANDDEIKITEDLSIVKTGTVEDDESADPRLAKDPAYEMFDKVDKFLQSHTLRVKVPEEITKSAASEYVPRSLLTDLPSELDMPLDGEDEAEVVEGRKKKIKLPKPLRIKSKHGFIKKVILPFLLGLKFKASVLVPLALALIALKTWKALTLGLISLVLSGAMVIFKFTKPKVVNYEVIHYPQHHVEHHVDHHAPAWDAHGPYQARSYEDAHEIAYSAQI
Summary
Uniprot
B6DXA9
I4DIU6
A0A194QJJ3
A0A0L7L6D8
A0A0N0PF22
A0A0L7LM80
+ More
I4DM28 A0A2H1V5U4 A0A212F9J6 A0A2W1BXJ2 A0A3S2NCF9 A0A1J1HHS0 B0WS68 Q17GL4 A0A336MLP6 A0A2J7PI78 A0A182F8I9 A0A067QXR6 A0A1B0DPT7 A0A182LVU6 A0A182YM63 A0A182VS82 A0A195DIX8 F4X728 A0A158NDB3 A0A182TYI5 A0A182HJX0 E2B2X6 A0A0C9R3U3 A0A182UZS5 A0A182KX57 A0A182XK21 Q7QDF4 A0A182R855 A0A182QDL1 A0A182PN18 A0A0L7RJ94 W5JEZ6 A0A195FHX5 A0A084WNS2 A0A182ITV8 E0VX97 A0A182NKZ3 A0A154PGZ4 A0A195B136 A0A1I8MW18 A0A151XHH7 A0A0A1XGV9 W8C8V4 A0A0M4EUL8 A0A0L0BQA7 A0A0K8WCB8 B4JYE2 A0A310SSF6 A0A034WD44 B4I4E2 A0A3B0JIP4 Q9VNP0 B3LVT6 A0A1I8PD93 B4QY98 A0A1W4URI4 B4PVP5 A0A0N0BK78 A0A195C1C7 B4KBN5 E2A508 A0A2A3EPZ9 B3NYN6 B4NHC0 A0A088AGG9 A0A3L8DFE1 Q298A3 A0A1B0FLA0 A0A1A9ZSX2 A0A232FAW9 J3JVV4 B4G3B4 A0A1A9W5P0 B4MBN9 A0A1A9YQ56 D6X0W6 A0A0J7KMV1 A0A1A9VDN1 N6UKM2 A0A1B0B2X0 K7IU87 A0A026WHU2 A0A2S2PW83 J9K7Y8 C4WSC5 A0A170Z9R9 T1HK55
I4DM28 A0A2H1V5U4 A0A212F9J6 A0A2W1BXJ2 A0A3S2NCF9 A0A1J1HHS0 B0WS68 Q17GL4 A0A336MLP6 A0A2J7PI78 A0A182F8I9 A0A067QXR6 A0A1B0DPT7 A0A182LVU6 A0A182YM63 A0A182VS82 A0A195DIX8 F4X728 A0A158NDB3 A0A182TYI5 A0A182HJX0 E2B2X6 A0A0C9R3U3 A0A182UZS5 A0A182KX57 A0A182XK21 Q7QDF4 A0A182R855 A0A182QDL1 A0A182PN18 A0A0L7RJ94 W5JEZ6 A0A195FHX5 A0A084WNS2 A0A182ITV8 E0VX97 A0A182NKZ3 A0A154PGZ4 A0A195B136 A0A1I8MW18 A0A151XHH7 A0A0A1XGV9 W8C8V4 A0A0M4EUL8 A0A0L0BQA7 A0A0K8WCB8 B4JYE2 A0A310SSF6 A0A034WD44 B4I4E2 A0A3B0JIP4 Q9VNP0 B3LVT6 A0A1I8PD93 B4QY98 A0A1W4URI4 B4PVP5 A0A0N0BK78 A0A195C1C7 B4KBN5 E2A508 A0A2A3EPZ9 B3NYN6 B4NHC0 A0A088AGG9 A0A3L8DFE1 Q298A3 A0A1B0FLA0 A0A1A9ZSX2 A0A232FAW9 J3JVV4 B4G3B4 A0A1A9W5P0 B4MBN9 A0A1A9YQ56 D6X0W6 A0A0J7KMV1 A0A1A9VDN1 N6UKM2 A0A1B0B2X0 K7IU87 A0A026WHU2 A0A2S2PW83 J9K7Y8 C4WSC5 A0A170Z9R9 T1HK55
Pubmed
19121390
22651552
26354079
26227816
22118469
28756777
+ More
17510324 24845553 25244985 21719571 21347285 20798317 20966253 12364791 14747013 17210077 20920257 23761445 24438588 20566863 25315136 25830018 24495485 26108605 17994087 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 30249741 15632085 28648823 22516182 18362917 19820115 23537049 20075255 24508170
17510324 24845553 25244985 21719571 21347285 20798317 20966253 12364791 14747013 17210077 20920257 23761445 24438588 20566863 25315136 25830018 24495485 26108605 17994087 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 30249741 15632085 28648823 22516182 18362917 19820115 23537049 20075255 24508170
EMBL
BABH01035651
FJ176297
ACI23619.1
AK401214
BAM17836.1
KQ459144
+ More
KPJ03621.1 JTDY01002673 KOB70970.1 KQ459717 KPJ20257.1 JTDY01000590 KOB76552.1 AK402346 BAM18968.1 ODYU01000846 SOQ36220.1 AGBW02009573 OWR50401.1 KZ149905 PZC78365.1 RSAL01000230 RVE43874.1 CVRI01000003 CRK87098.1 DS232065 EDS33691.1 CH477258 EAT45785.1 UFQT01001421 SSX30481.1 NEVH01025129 PNF16046.1 KK852895 KDR14185.1 AJVK01018563 AXCM01009100 KQ980800 KYN12843.1 GL888828 EGI57640.1 ADTU01012542 APCN01002211 GL445250 EFN89945.1 GBYB01007467 JAG77234.1 AAAB01008851 EAA07339.5 AXCN02000107 KQ414581 KOC70935.1 ADMH02001348 ETN62932.1 KQ981560 KYN39961.1 ATLV01024674 KE525356 KFB51866.1 DS235830 EEB18003.1 KQ434902 KZC11129.1 KQ976690 KYM78005.1 KQ982130 KYQ59807.1 GBXI01004100 JAD10192.1 GAMC01002719 JAC03837.1 CP012526 ALC46817.1 JRES01001623 KNC21379.1 GDHF01003491 JAI48823.1 CH916377 EDV90704.1 KQ759821 OAD62717.1 GAKP01005441 JAC53511.1 CH480821 EDW55085.1 OUUW01000005 SPP80192.1 AE014297 AY070982 AAF51889.2 AAL48604.1 CH902617 EDV43710.1 CM000364 EDX11855.1 CM000160 EDW97854.1 KQ435706 KOX80184.1 KQ978379 KYM94420.1 CH933806 EDW14712.1 GL436778 EFN71495.1 KZ288200 PBC33584.1 CH954181 EDV48017.1 CH964272 EDW84596.1 QOIP01000009 RLU18619.1 CM000070 EAL28052.2 CCAG010013281 NNAY01000498 OXU27986.1 BT127372 AEE62334.1 CH479179 EDW24296.1 CH940656 EDW58510.1 KQ971372 EFA10564.1 LBMM01005384 KMQ91564.1 APGK01029598 APGK01029599 KB740735 KB631809 ENN79232.1 ERL86323.1 JXJN01007737 AAZX01004766 AAZX01006235 KK107199 EZA55602.1 GGMS01000460 MBY69663.1 ABLF02034092 AK340137 BAH70795.1 GEMB01002456 JAS00730.1 ACPB03024080
KPJ03621.1 JTDY01002673 KOB70970.1 KQ459717 KPJ20257.1 JTDY01000590 KOB76552.1 AK402346 BAM18968.1 ODYU01000846 SOQ36220.1 AGBW02009573 OWR50401.1 KZ149905 PZC78365.1 RSAL01000230 RVE43874.1 CVRI01000003 CRK87098.1 DS232065 EDS33691.1 CH477258 EAT45785.1 UFQT01001421 SSX30481.1 NEVH01025129 PNF16046.1 KK852895 KDR14185.1 AJVK01018563 AXCM01009100 KQ980800 KYN12843.1 GL888828 EGI57640.1 ADTU01012542 APCN01002211 GL445250 EFN89945.1 GBYB01007467 JAG77234.1 AAAB01008851 EAA07339.5 AXCN02000107 KQ414581 KOC70935.1 ADMH02001348 ETN62932.1 KQ981560 KYN39961.1 ATLV01024674 KE525356 KFB51866.1 DS235830 EEB18003.1 KQ434902 KZC11129.1 KQ976690 KYM78005.1 KQ982130 KYQ59807.1 GBXI01004100 JAD10192.1 GAMC01002719 JAC03837.1 CP012526 ALC46817.1 JRES01001623 KNC21379.1 GDHF01003491 JAI48823.1 CH916377 EDV90704.1 KQ759821 OAD62717.1 GAKP01005441 JAC53511.1 CH480821 EDW55085.1 OUUW01000005 SPP80192.1 AE014297 AY070982 AAF51889.2 AAL48604.1 CH902617 EDV43710.1 CM000364 EDX11855.1 CM000160 EDW97854.1 KQ435706 KOX80184.1 KQ978379 KYM94420.1 CH933806 EDW14712.1 GL436778 EFN71495.1 KZ288200 PBC33584.1 CH954181 EDV48017.1 CH964272 EDW84596.1 QOIP01000009 RLU18619.1 CM000070 EAL28052.2 CCAG010013281 NNAY01000498 OXU27986.1 BT127372 AEE62334.1 CH479179 EDW24296.1 CH940656 EDW58510.1 KQ971372 EFA10564.1 LBMM01005384 KMQ91564.1 APGK01029598 APGK01029599 KB740735 KB631809 ENN79232.1 ERL86323.1 JXJN01007737 AAZX01004766 AAZX01006235 KK107199 EZA55602.1 GGMS01000460 MBY69663.1 ABLF02034092 AK340137 BAH70795.1 GEMB01002456 JAS00730.1 ACPB03024080
Proteomes
UP000005204
UP000053268
UP000037510
UP000053240
UP000007151
UP000283053
+ More
UP000183832 UP000002320 UP000008820 UP000235965 UP000069272 UP000027135 UP000092462 UP000075883 UP000076408 UP000075920 UP000078492 UP000007755 UP000005205 UP000075902 UP000075840 UP000008237 UP000075903 UP000075882 UP000076407 UP000007062 UP000075900 UP000075886 UP000075885 UP000053825 UP000000673 UP000078541 UP000030765 UP000075880 UP000009046 UP000075884 UP000076502 UP000078540 UP000095301 UP000075809 UP000092553 UP000037069 UP000001070 UP000001292 UP000268350 UP000000803 UP000007801 UP000095300 UP000000304 UP000192221 UP000002282 UP000053105 UP000078542 UP000009192 UP000000311 UP000242457 UP000008711 UP000007798 UP000005203 UP000279307 UP000001819 UP000092444 UP000092445 UP000215335 UP000008744 UP000091820 UP000008792 UP000092443 UP000007266 UP000036403 UP000078200 UP000019118 UP000030742 UP000092460 UP000002358 UP000053097 UP000007819 UP000015103
UP000183832 UP000002320 UP000008820 UP000235965 UP000069272 UP000027135 UP000092462 UP000075883 UP000076408 UP000075920 UP000078492 UP000007755 UP000005205 UP000075902 UP000075840 UP000008237 UP000075903 UP000075882 UP000076407 UP000007062 UP000075900 UP000075886 UP000075885 UP000053825 UP000000673 UP000078541 UP000030765 UP000075880 UP000009046 UP000075884 UP000076502 UP000078540 UP000095301 UP000075809 UP000092553 UP000037069 UP000001070 UP000001292 UP000268350 UP000000803 UP000007801 UP000095300 UP000000304 UP000192221 UP000002282 UP000053105 UP000078542 UP000009192 UP000000311 UP000242457 UP000008711 UP000007798 UP000005203 UP000279307 UP000001819 UP000092444 UP000092445 UP000215335 UP000008744 UP000091820 UP000008792 UP000092443 UP000007266 UP000036403 UP000078200 UP000019118 UP000030742 UP000092460 UP000002358 UP000053097 UP000007819 UP000015103
Pfam
PF07898 DUF1676
Interpro
IPR012464
DUF1676
ProteinModelPortal
B6DXA9
I4DIU6
A0A194QJJ3
A0A0L7L6D8
A0A0N0PF22
A0A0L7LM80
+ More
I4DM28 A0A2H1V5U4 A0A212F9J6 A0A2W1BXJ2 A0A3S2NCF9 A0A1J1HHS0 B0WS68 Q17GL4 A0A336MLP6 A0A2J7PI78 A0A182F8I9 A0A067QXR6 A0A1B0DPT7 A0A182LVU6 A0A182YM63 A0A182VS82 A0A195DIX8 F4X728 A0A158NDB3 A0A182TYI5 A0A182HJX0 E2B2X6 A0A0C9R3U3 A0A182UZS5 A0A182KX57 A0A182XK21 Q7QDF4 A0A182R855 A0A182QDL1 A0A182PN18 A0A0L7RJ94 W5JEZ6 A0A195FHX5 A0A084WNS2 A0A182ITV8 E0VX97 A0A182NKZ3 A0A154PGZ4 A0A195B136 A0A1I8MW18 A0A151XHH7 A0A0A1XGV9 W8C8V4 A0A0M4EUL8 A0A0L0BQA7 A0A0K8WCB8 B4JYE2 A0A310SSF6 A0A034WD44 B4I4E2 A0A3B0JIP4 Q9VNP0 B3LVT6 A0A1I8PD93 B4QY98 A0A1W4URI4 B4PVP5 A0A0N0BK78 A0A195C1C7 B4KBN5 E2A508 A0A2A3EPZ9 B3NYN6 B4NHC0 A0A088AGG9 A0A3L8DFE1 Q298A3 A0A1B0FLA0 A0A1A9ZSX2 A0A232FAW9 J3JVV4 B4G3B4 A0A1A9W5P0 B4MBN9 A0A1A9YQ56 D6X0W6 A0A0J7KMV1 A0A1A9VDN1 N6UKM2 A0A1B0B2X0 K7IU87 A0A026WHU2 A0A2S2PW83 J9K7Y8 C4WSC5 A0A170Z9R9 T1HK55
I4DM28 A0A2H1V5U4 A0A212F9J6 A0A2W1BXJ2 A0A3S2NCF9 A0A1J1HHS0 B0WS68 Q17GL4 A0A336MLP6 A0A2J7PI78 A0A182F8I9 A0A067QXR6 A0A1B0DPT7 A0A182LVU6 A0A182YM63 A0A182VS82 A0A195DIX8 F4X728 A0A158NDB3 A0A182TYI5 A0A182HJX0 E2B2X6 A0A0C9R3U3 A0A182UZS5 A0A182KX57 A0A182XK21 Q7QDF4 A0A182R855 A0A182QDL1 A0A182PN18 A0A0L7RJ94 W5JEZ6 A0A195FHX5 A0A084WNS2 A0A182ITV8 E0VX97 A0A182NKZ3 A0A154PGZ4 A0A195B136 A0A1I8MW18 A0A151XHH7 A0A0A1XGV9 W8C8V4 A0A0M4EUL8 A0A0L0BQA7 A0A0K8WCB8 B4JYE2 A0A310SSF6 A0A034WD44 B4I4E2 A0A3B0JIP4 Q9VNP0 B3LVT6 A0A1I8PD93 B4QY98 A0A1W4URI4 B4PVP5 A0A0N0BK78 A0A195C1C7 B4KBN5 E2A508 A0A2A3EPZ9 B3NYN6 B4NHC0 A0A088AGG9 A0A3L8DFE1 Q298A3 A0A1B0FLA0 A0A1A9ZSX2 A0A232FAW9 J3JVV4 B4G3B4 A0A1A9W5P0 B4MBN9 A0A1A9YQ56 D6X0W6 A0A0J7KMV1 A0A1A9VDN1 N6UKM2 A0A1B0B2X0 K7IU87 A0A026WHU2 A0A2S2PW83 J9K7Y8 C4WSC5 A0A170Z9R9 T1HK55
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.990338
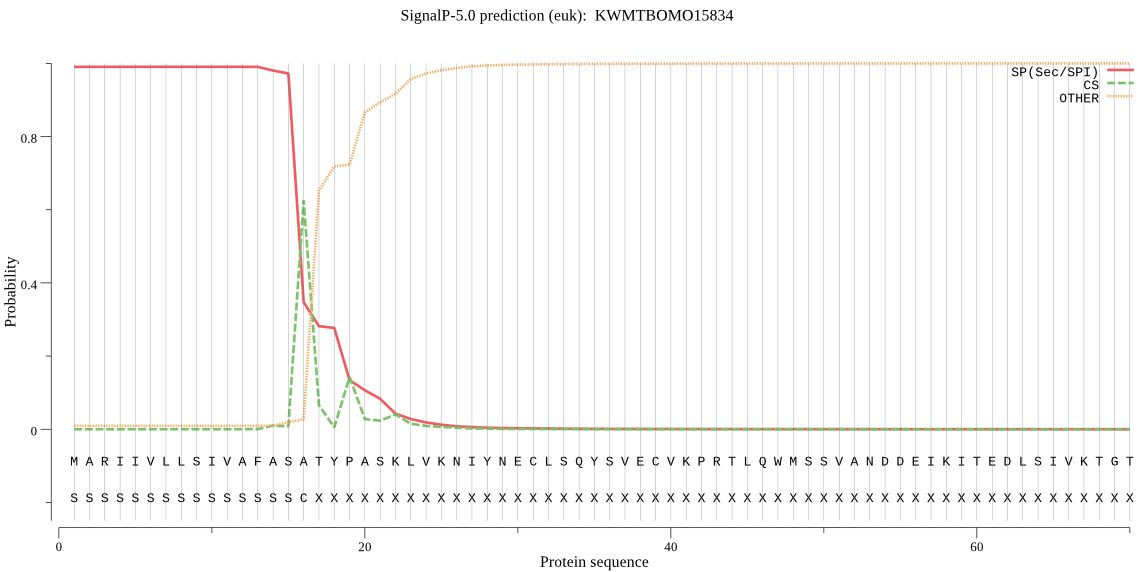
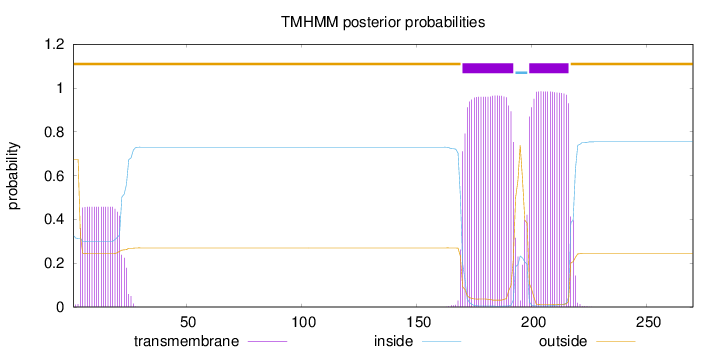
Length:
270
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
50.42349
Exp number, first 60 AAs:
8.85324
Total prob of N-in:
0.32587
outside
1 - 169
TMhelix
170 - 192
inside
193 - 198
TMhelix
199 - 216
outside
217 - 270
Population Genetic Test Statistics
Pi
137.562853
Theta
119.285372
Tajima's D
0.367262
CLR
9.481317
CSRT
0.464376781160942
Interpretation
Uncertain
