Gene
KWMTBOMO15825
Pre Gene Modal
BGIBMGA000047
Annotation
PREDICTED:_uncharacterized_protein_LOC106130656_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.693
Sequence
CDS
ATGTACACAGTGCTCCTTCTAACCGTCCTTCTTACTACCACATTTGCTAATGAAGACCTCAGCTATAAATACCTCACGAAAGTATACGATGAATGCCAGAAATCGGACGCGATTTTGCCATGTTTGAAGAAGAAAGCTATATTGTTCTTCGATCGGGCCGCCAGGATGGAAACGATACCTTTGCTAGACGGAGTGGACATTGTGAAGGTGTCAAAGACAGATGAAAACGTTCCCATAAGCGAGAATGAGATAGACGCGGCACTCCCCAGGAATTTGCAGGATAAAAATGAAGCCTTAACGCAAATGCTGTGGGATAAAGTGGCGAGTTTCGCTAATTCGAGAACAATACAACTAGCCTTACCGAAGATGACTGGCGCAGAATTAAACAGAGGTGTTGAAGAAGGTAAGTTTTATTTACTTCAAAGCGTAACGTTAGTATAA
Protein
MYTVLLLTVLLTTTFANEDLSYKYLTKVYDECQKSDAILPCLKKKAILFFDRAARMETIPLLDGVDIVKVSKTDENVPISENEIDAALPRNLQDKNEALTQMLWDKVASFANSRTIQLALPKMTGAELNRGVEEGKFYLLQSVTLV
Summary
Uniprot
H9IS21
A0A3S2LCP9
A0A0L7K420
A0A0L7KY24
A0A194QDK4
A0A2H1V0U7
+ More
A0A0N1PHR7 A0A2W1BXI3 A0A1J1HGW1 A0A182JZ29 W5JWL7 A0A182RYK0 A0A182F6Q3 A0A182Q9D5 A0A182PF67 A0A182YGJ5 A0A0L7RJ99 A0A182VE87 A0A154PH01 A0A182XIZ7 A0A182L7G1 F5HL35 A0A182I7W0 A0A182TRQ2 B3LVU5 A0A182MM28 N6TFR7 B0WLV4 A0A182JFX5 A0A182N5P6 A0A084VLA2 A0A182VSD0 A0A1W4V3R1 A0A182GF05 B4MBP8 A0A182GBT0 B4PVN2 B3NYM6 A0A1I8N768 Q297Z6 B4G3A5 Q9VNN2 B4I4D2 B4QY89 B4JYF0 T1GZ42 A0A0L0BN13 Q17GM1 A0A1I8QCL1 A0A336L843 B4KBM6 A0A088AGG1 E9IVC5 A0A1B0DBB3 A0A2P8YY82 A0A2A3EPG1 A0A1B0GHW5 A0A034WMS4 A0A0N0U7A1 A0A3L8DE94 E2B2X0 A0A3B0JHM2 B4MHL8 D6X0T7 A0A0K8UYK2 A0A151XHB6 B4NHC6 A0A0K8S8H4 A0A0M5J298 A0A0A1WVZ0 A0A195FHV0 A0A195DJ55 A0A195C2I7 A0A067RB11 A0A158NDH3 A0A0C9Q0F3 A0A0J7KMH2 A0A1W4WNB0 A0A2J7QXI9 N6TFR3 T1HNE1 E2API4 A0A0P4VIE5 R4G3B1 A0A146LVA2 A0A0A9XFU0 K7IU82 A0A2S2R4H0 D6X0U2 A0A158NDA8 A0A195B150 A0A195FHU4 A0A195DJ60 A0A1A9W5M9 F4X740 A0A026WKG1 A0A151XHB1
A0A0N1PHR7 A0A2W1BXI3 A0A1J1HGW1 A0A182JZ29 W5JWL7 A0A182RYK0 A0A182F6Q3 A0A182Q9D5 A0A182PF67 A0A182YGJ5 A0A0L7RJ99 A0A182VE87 A0A154PH01 A0A182XIZ7 A0A182L7G1 F5HL35 A0A182I7W0 A0A182TRQ2 B3LVU5 A0A182MM28 N6TFR7 B0WLV4 A0A182JFX5 A0A182N5P6 A0A084VLA2 A0A182VSD0 A0A1W4V3R1 A0A182GF05 B4MBP8 A0A182GBT0 B4PVN2 B3NYM6 A0A1I8N768 Q297Z6 B4G3A5 Q9VNN2 B4I4D2 B4QY89 B4JYF0 T1GZ42 A0A0L0BN13 Q17GM1 A0A1I8QCL1 A0A336L843 B4KBM6 A0A088AGG1 E9IVC5 A0A1B0DBB3 A0A2P8YY82 A0A2A3EPG1 A0A1B0GHW5 A0A034WMS4 A0A0N0U7A1 A0A3L8DE94 E2B2X0 A0A3B0JHM2 B4MHL8 D6X0T7 A0A0K8UYK2 A0A151XHB6 B4NHC6 A0A0K8S8H4 A0A0M5J298 A0A0A1WVZ0 A0A195FHV0 A0A195DJ55 A0A195C2I7 A0A067RB11 A0A158NDH3 A0A0C9Q0F3 A0A0J7KMH2 A0A1W4WNB0 A0A2J7QXI9 N6TFR3 T1HNE1 E2API4 A0A0P4VIE5 R4G3B1 A0A146LVA2 A0A0A9XFU0 K7IU82 A0A2S2R4H0 D6X0U2 A0A158NDA8 A0A195B150 A0A195FHU4 A0A195DJ60 A0A1A9W5M9 F4X740 A0A026WKG1 A0A151XHB1
Pubmed
19121390
26227816
26354079
28756777
20920257
23761445
+ More
25244985 20966253 12364791 14747013 17210077 17994087 23537049 24438588 26483478 17550304 25315136 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 17510324 21282665 29403074 25348373 30249741 20798317 18362917 19820115 25830018 24845553 21347285 27129103 26823975 25401762 20075255 21719571 24508170
25244985 20966253 12364791 14747013 17210077 17994087 23537049 24438588 26483478 17550304 25315136 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 17510324 21282665 29403074 25348373 30249741 20798317 18362917 19820115 25830018 24845553 21347285 27129103 26823975 25401762 20075255 21719571 24508170
EMBL
BABH01035642
RSAL01000230
RVE43864.1
JTDY01011676
KOB54455.1
JTDY01004615
+ More
KOB67944.1 KQ459144 KPJ03628.1 ODYU01000169 SOQ34483.1 KQ459717 KPJ20264.1 KZ149905 PZC78355.1 CVRI01000003 CRK87115.1 ADMH02000198 ETN67394.1 AXCN02001159 KQ414581 KOC70940.1 KQ434902 KZC11135.1 AAAB01008880 EGK96996.1 APCN01002561 CH902617 EDV43719.1 AXCM01004200 APGK01029591 KB740735 KB631809 ENN79224.1 ERL86315.1 DS231991 EDS30689.1 ATLV01014437 KE524972 KFB38746.1 JXUM01059331 KQ562047 KXJ76817.1 CH940656 EDW58519.2 JXUM01054115 KQ561806 KXJ77474.1 CM000160 EDW97841.1 CH954181 EDV48007.1 CM000070 EAL28059.2 CH479179 EDW24287.1 AE014297 BT009951 AAF51897.2 AAQ22420.1 CH480821 EDW55075.1 CM000364 EDX11846.1 CH916377 EDV90712.1 CAQQ02178678 JRES01001623 KNC21377.1 CH477258 EAT45778.1 UFQS01002245 UFQT01002223 UFQT01002245 SSX13589.1 SSX32962.1 CH933806 EDW14703.1 GL766194 EFZ15484.1 AJVK01013610 PYGN01000289 PSN49209.1 KZ288200 PBC33590.1 AJWK01005741 GAKP01003477 JAC55475.1 KQ435706 KOX80178.1 QOIP01000009 RLU18755.1 GL445250 EFN89939.1 OUUW01000005 SPP80203.1 CH959795 EDW71607.2 KQ971372 EFA09555.1 GDHF01020618 JAI31696.1 KQ982130 KYQ59799.1 CH964272 EDW84602.2 GBRD01016249 JAG49577.1 CP012526 ALC46824.1 GBXI01011270 JAD03022.1 KQ981560 KYN39953.1 KQ980800 KYN12852.1 KQ978379 KYM94426.1 KK852623 KDR19954.1 ADTU01012537 GBYB01007303 JAG77070.1 LBMM01005451 KMQ91502.1 NEVH01009379 PNF33301.1 APGK01029589 ENN79219.1 ERL86310.1 ACPB03007640 GL441542 EFN64640.1 GDKW01002956 JAI53639.1 GAHY01002355 JAA75155.1 GDHC01008339 JAQ10290.1 GBHO01025966 JAG17638.1 AAZX01004766 GGMS01015665 MBY84868.1 EFA09551.1 KQ976690 KYM78020.1 KYN39948.1 KYN12857.1 GL888828 EGI57652.1 KK107199 EZA55589.1 KYQ59794.1
KOB67944.1 KQ459144 KPJ03628.1 ODYU01000169 SOQ34483.1 KQ459717 KPJ20264.1 KZ149905 PZC78355.1 CVRI01000003 CRK87115.1 ADMH02000198 ETN67394.1 AXCN02001159 KQ414581 KOC70940.1 KQ434902 KZC11135.1 AAAB01008880 EGK96996.1 APCN01002561 CH902617 EDV43719.1 AXCM01004200 APGK01029591 KB740735 KB631809 ENN79224.1 ERL86315.1 DS231991 EDS30689.1 ATLV01014437 KE524972 KFB38746.1 JXUM01059331 KQ562047 KXJ76817.1 CH940656 EDW58519.2 JXUM01054115 KQ561806 KXJ77474.1 CM000160 EDW97841.1 CH954181 EDV48007.1 CM000070 EAL28059.2 CH479179 EDW24287.1 AE014297 BT009951 AAF51897.2 AAQ22420.1 CH480821 EDW55075.1 CM000364 EDX11846.1 CH916377 EDV90712.1 CAQQ02178678 JRES01001623 KNC21377.1 CH477258 EAT45778.1 UFQS01002245 UFQT01002223 UFQT01002245 SSX13589.1 SSX32962.1 CH933806 EDW14703.1 GL766194 EFZ15484.1 AJVK01013610 PYGN01000289 PSN49209.1 KZ288200 PBC33590.1 AJWK01005741 GAKP01003477 JAC55475.1 KQ435706 KOX80178.1 QOIP01000009 RLU18755.1 GL445250 EFN89939.1 OUUW01000005 SPP80203.1 CH959795 EDW71607.2 KQ971372 EFA09555.1 GDHF01020618 JAI31696.1 KQ982130 KYQ59799.1 CH964272 EDW84602.2 GBRD01016249 JAG49577.1 CP012526 ALC46824.1 GBXI01011270 JAD03022.1 KQ981560 KYN39953.1 KQ980800 KYN12852.1 KQ978379 KYM94426.1 KK852623 KDR19954.1 ADTU01012537 GBYB01007303 JAG77070.1 LBMM01005451 KMQ91502.1 NEVH01009379 PNF33301.1 APGK01029589 ENN79219.1 ERL86310.1 ACPB03007640 GL441542 EFN64640.1 GDKW01002956 JAI53639.1 GAHY01002355 JAA75155.1 GDHC01008339 JAQ10290.1 GBHO01025966 JAG17638.1 AAZX01004766 GGMS01015665 MBY84868.1 EFA09551.1 KQ976690 KYM78020.1 KYN39948.1 KYN12857.1 GL888828 EGI57652.1 KK107199 EZA55589.1 KYQ59794.1
Proteomes
UP000005204
UP000283053
UP000037510
UP000053268
UP000053240
UP000183832
+ More
UP000075881 UP000000673 UP000075900 UP000069272 UP000075886 UP000075885 UP000076408 UP000053825 UP000075903 UP000076502 UP000076407 UP000075882 UP000007062 UP000075840 UP000075902 UP000007801 UP000075883 UP000019118 UP000030742 UP000002320 UP000075880 UP000075884 UP000030765 UP000075920 UP000192221 UP000069940 UP000249989 UP000008792 UP000002282 UP000008711 UP000095301 UP000001819 UP000008744 UP000000803 UP000001292 UP000000304 UP000001070 UP000015102 UP000037069 UP000008820 UP000095300 UP000009192 UP000005203 UP000092462 UP000245037 UP000242457 UP000092461 UP000053105 UP000279307 UP000008237 UP000268350 UP000007798 UP000007266 UP000075809 UP000092553 UP000078541 UP000078492 UP000078542 UP000027135 UP000005205 UP000036403 UP000192223 UP000235965 UP000015103 UP000000311 UP000002358 UP000078540 UP000091820 UP000007755 UP000053097
UP000075881 UP000000673 UP000075900 UP000069272 UP000075886 UP000075885 UP000076408 UP000053825 UP000075903 UP000076502 UP000076407 UP000075882 UP000007062 UP000075840 UP000075902 UP000007801 UP000075883 UP000019118 UP000030742 UP000002320 UP000075880 UP000075884 UP000030765 UP000075920 UP000192221 UP000069940 UP000249989 UP000008792 UP000002282 UP000008711 UP000095301 UP000001819 UP000008744 UP000000803 UP000001292 UP000000304 UP000001070 UP000015102 UP000037069 UP000008820 UP000095300 UP000009192 UP000005203 UP000092462 UP000245037 UP000242457 UP000092461 UP000053105 UP000279307 UP000008237 UP000268350 UP000007798 UP000007266 UP000075809 UP000092553 UP000078541 UP000078492 UP000078542 UP000027135 UP000005205 UP000036403 UP000192223 UP000235965 UP000015103 UP000000311 UP000002358 UP000078540 UP000091820 UP000007755 UP000053097
Interpro
SUPFAM
SSF54593
SSF54593
Gene 3D
ProteinModelPortal
H9IS21
A0A3S2LCP9
A0A0L7K420
A0A0L7KY24
A0A194QDK4
A0A2H1V0U7
+ More
A0A0N1PHR7 A0A2W1BXI3 A0A1J1HGW1 A0A182JZ29 W5JWL7 A0A182RYK0 A0A182F6Q3 A0A182Q9D5 A0A182PF67 A0A182YGJ5 A0A0L7RJ99 A0A182VE87 A0A154PH01 A0A182XIZ7 A0A182L7G1 F5HL35 A0A182I7W0 A0A182TRQ2 B3LVU5 A0A182MM28 N6TFR7 B0WLV4 A0A182JFX5 A0A182N5P6 A0A084VLA2 A0A182VSD0 A0A1W4V3R1 A0A182GF05 B4MBP8 A0A182GBT0 B4PVN2 B3NYM6 A0A1I8N768 Q297Z6 B4G3A5 Q9VNN2 B4I4D2 B4QY89 B4JYF0 T1GZ42 A0A0L0BN13 Q17GM1 A0A1I8QCL1 A0A336L843 B4KBM6 A0A088AGG1 E9IVC5 A0A1B0DBB3 A0A2P8YY82 A0A2A3EPG1 A0A1B0GHW5 A0A034WMS4 A0A0N0U7A1 A0A3L8DE94 E2B2X0 A0A3B0JHM2 B4MHL8 D6X0T7 A0A0K8UYK2 A0A151XHB6 B4NHC6 A0A0K8S8H4 A0A0M5J298 A0A0A1WVZ0 A0A195FHV0 A0A195DJ55 A0A195C2I7 A0A067RB11 A0A158NDH3 A0A0C9Q0F3 A0A0J7KMH2 A0A1W4WNB0 A0A2J7QXI9 N6TFR3 T1HNE1 E2API4 A0A0P4VIE5 R4G3B1 A0A146LVA2 A0A0A9XFU0 K7IU82 A0A2S2R4H0 D6X0U2 A0A158NDA8 A0A195B150 A0A195FHU4 A0A195DJ60 A0A1A9W5M9 F4X740 A0A026WKG1 A0A151XHB1
A0A0N1PHR7 A0A2W1BXI3 A0A1J1HGW1 A0A182JZ29 W5JWL7 A0A182RYK0 A0A182F6Q3 A0A182Q9D5 A0A182PF67 A0A182YGJ5 A0A0L7RJ99 A0A182VE87 A0A154PH01 A0A182XIZ7 A0A182L7G1 F5HL35 A0A182I7W0 A0A182TRQ2 B3LVU5 A0A182MM28 N6TFR7 B0WLV4 A0A182JFX5 A0A182N5P6 A0A084VLA2 A0A182VSD0 A0A1W4V3R1 A0A182GF05 B4MBP8 A0A182GBT0 B4PVN2 B3NYM6 A0A1I8N768 Q297Z6 B4G3A5 Q9VNN2 B4I4D2 B4QY89 B4JYF0 T1GZ42 A0A0L0BN13 Q17GM1 A0A1I8QCL1 A0A336L843 B4KBM6 A0A088AGG1 E9IVC5 A0A1B0DBB3 A0A2P8YY82 A0A2A3EPG1 A0A1B0GHW5 A0A034WMS4 A0A0N0U7A1 A0A3L8DE94 E2B2X0 A0A3B0JHM2 B4MHL8 D6X0T7 A0A0K8UYK2 A0A151XHB6 B4NHC6 A0A0K8S8H4 A0A0M5J298 A0A0A1WVZ0 A0A195FHV0 A0A195DJ55 A0A195C2I7 A0A067RB11 A0A158NDH3 A0A0C9Q0F3 A0A0J7KMH2 A0A1W4WNB0 A0A2J7QXI9 N6TFR3 T1HNE1 E2API4 A0A0P4VIE5 R4G3B1 A0A146LVA2 A0A0A9XFU0 K7IU82 A0A2S2R4H0 D6X0U2 A0A158NDA8 A0A195B150 A0A195FHU4 A0A195DJ60 A0A1A9W5M9 F4X740 A0A026WKG1 A0A151XHB1
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.993168
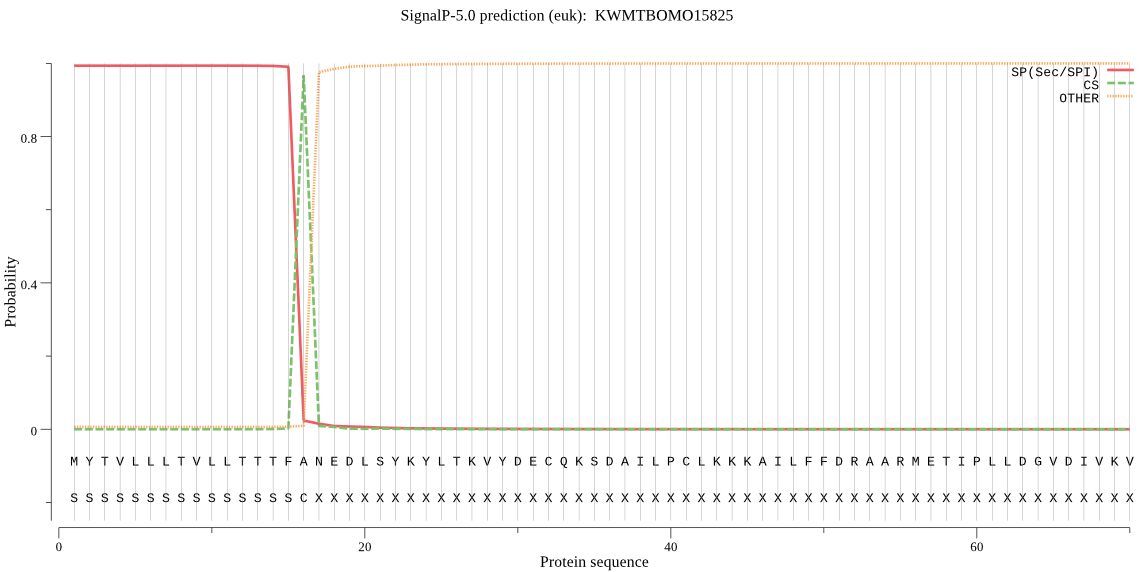
Length:
146
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06163
Exp number, first 60 AAs:
0.06163
Total prob of N-in:
0.03984
outside
1 - 146

Population Genetic Test Statistics
Pi
269.709194
Theta
215.110059
Tajima's D
0.79916
CLR
0.011521
CSRT
0.600769961501925
Interpretation
Uncertain
