Pre Gene Modal
BGIBMGA000008
Annotation
PREDICTED:_uncharacterized_protein_LOC106130657_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.26
Sequence
CDS
ATGCTGTGCCGCGGGTTCGTCCTCTGCCTCGCCGTAGCAGCCGTCACCGCTGACACCCTCACCAGCTTCAAGATCCTAGATAAAATAAGTGGACAGTGCGGGAAATACCCTTCAAAGGATATCCTCACTTGTTTTAAGTTCCAAGCTGCTAAGTTCGTAGATCGAGCGGCCAGATTGGAGTCTCTACCGATTGCTGATGGCGTCACCTTAGTACGGAGACCGGAAGCGGAAAGAGCGTTTCCCTCTTCATCGCCGGAAGGAGATTTAAATTCTTTATCATCTGACCAAGTGGACAAGTATTTACAGGATGCGACCTCGAAGTTGATGCAGACGCATCGGGTGGTCATTGCCCCTCAGACCTTTGGGGAAGACGTTGGTAGATCTATCAATGAAGCTAGAGGCAAGCTGAAGAAGATGATGGGGCCAATTATCGCTGGTCTTGCGATCAAGGGAGGCTTCGTAGCGATTGCTTTTCAAGCAATAGCTCTTATTGCTGGAAAGGCGCTGCTCATAGGAAAAATTGCTCTGCTGCTGTCAGCCATTATCGGCTTGAAGAAGCTAGTCGCCGGAGGAGACACCCACGAGAAGACGACGTATGAGATCGTTAAGCATCCGCAGGTATCTCAAAGTCATACGTACTCTTCTTCCCATTACGGCGGTGATTTCGACAGCACCGGTCCAGGAGGACACTATAGGAGGAGCGTGGAAGATGAGGCAGCAGCCCAAGATAGAGCGTATAGGGCCTATGCCACGAAGCAGTAG
Protein
MLCRGFVLCLAVAAVTADTLTSFKILDKISGQCGKYPSKDILTCFKFQAAKFVDRAARLESLPIADGVTLVRRPEAERAFPSSSPEGDLNSLSSDQVDKYLQDATSKLMQTHRVVIAPQTFGEDVGRSINEARGKLKKMMGPIIAGLAIKGGFVAIAFQAIALIAGKALLIGKIALLLSAIIGLKKLVAGGDTHEKTTYEIVKHPQVSQSHTYSSSHYGGDFDSTGPGGHYRRSVEDEAAAQDRAYRAYATKQ
Summary
Uniprot
H9IRY8
A0A0N1IH62
A0A0L7KXE4
A0A194QDY0
A0A2W1BXJ0
A0A2H1V2I8
+ More
A0A212EVS1 B0WLV5 A0A084VL98 A0A182GBT1 A0A1S4F7C9 Q17DB0 A0A182IN33 A0A182F6Q2 W5JL05 A0A182VSC9 A0A182MTD6 A0A182GF06 A0A182YGJ4 A0A2P8YY80 A0A182PF66 A0A182VAW6 A0A182THY0 A0A182XIZ6 A0A182L7G8 A7USY6 A0A182I7W1 A0A182RYJ9 A0A182N5P5 A0A182QTI9 A0A2R7VU74 A0A1J1HGH6 T1HNE3 B4MBP9 A0A1W4URM9 B3LVU6 B4QY88 B3NYM5 B4PVN1 A0A1B0DBB2 B4I4D1 A0A0L0BMR2 B4JYF1 Q9VNN1 B4KBM5 A0A0M4F4T2 A0A1B0GHW4 B4G3A4 A0A1I8PD50 A0A336L7Z4 Q297Z5 A0A2P8YY81 A0A3B0JDQ7 A0A0Q9WNG2 A0A0K8U631 N6TMG8 B4NHD1 A0A3B0JCU6 E0VA41 A0A1B0FJ43 A0A1A9ZSX7 A0A1I8MJ29 A0A034WEG0 A0A1B0B2Y0 A0A1A9YQ75 A0A1A9VDN7 A0A067R8N4 A0A0A1X5R7 U4TXI5 D6X0T8 A0A2J7QXJ5 A0A2S2R4H0 A0A1A9W5N3 A0A2S2PSH4 I4DLU7 J9JS56 A0A0N1PJF6 I4DID7
A0A212EVS1 B0WLV5 A0A084VL98 A0A182GBT1 A0A1S4F7C9 Q17DB0 A0A182IN33 A0A182F6Q2 W5JL05 A0A182VSC9 A0A182MTD6 A0A182GF06 A0A182YGJ4 A0A2P8YY80 A0A182PF66 A0A182VAW6 A0A182THY0 A0A182XIZ6 A0A182L7G8 A7USY6 A0A182I7W1 A0A182RYJ9 A0A182N5P5 A0A182QTI9 A0A2R7VU74 A0A1J1HGH6 T1HNE3 B4MBP9 A0A1W4URM9 B3LVU6 B4QY88 B3NYM5 B4PVN1 A0A1B0DBB2 B4I4D1 A0A0L0BMR2 B4JYF1 Q9VNN1 B4KBM5 A0A0M4F4T2 A0A1B0GHW4 B4G3A4 A0A1I8PD50 A0A336L7Z4 Q297Z5 A0A2P8YY81 A0A3B0JDQ7 A0A0Q9WNG2 A0A0K8U631 N6TMG8 B4NHD1 A0A3B0JCU6 E0VA41 A0A1B0FJ43 A0A1A9ZSX7 A0A1I8MJ29 A0A034WEG0 A0A1B0B2Y0 A0A1A9YQ75 A0A1A9VDN7 A0A067R8N4 A0A0A1X5R7 U4TXI5 D6X0T8 A0A2J7QXJ5 A0A2S2R4H0 A0A1A9W5N3 A0A2S2PSH4 I4DLU7 J9JS56 A0A0N1PJF6 I4DID7
Pubmed
19121390
26354079
26227816
28756777
22118469
24438588
+ More
26483478 17510324 20920257 23761445 25244985 29403074 20966253 12364791 14747013 17210077 17994087 17550304 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23537049 20566863 25315136 25348373 24845553 25830018 18362917 19820115 22651552
26483478 17510324 20920257 23761445 25244985 29403074 20966253 12364791 14747013 17210077 17994087 17550304 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23537049 20566863 25315136 25348373 24845553 25830018 18362917 19820115 22651552
EMBL
BABH01035641
KQ459717
KPJ20265.1
JTDY01004615
KOB67943.1
KQ459144
+ More
KPJ03629.1 KZ149905 PZC78354.1 ODYU01000169 SOQ34484.1 AGBW02012152 OWR45561.1 DS231991 EDS30690.1 ATLV01014432 KE524972 KFB38742.1 JXUM01054123 KQ561806 KXJ77475.1 CH477298 EAT44319.1 ADMH02000786 ETN65057.1 AXCM01010351 JXUM01059338 JXUM01059339 PYGN01000289 PSN49208.1 AAAB01008880 EDO64173.2 APCN01002561 AXCN02001159 KK854089 PTY11054.1 CVRI01000003 CRK87120.1 ACPB03007640 CH940656 EDW58520.1 CH902617 EDV43720.1 CM000364 EDX11845.1 CH954181 EDV48006.1 CM000160 EDW97840.1 AJVK01013609 CH480821 EDW55074.1 JRES01001623 KNC21375.1 CH916377 EDV90713.1 AE014297 BT100070 AAF51898.1 ACX61611.1 CH933806 EDW14702.1 CP012526 ALC46825.1 AJWK01005739 CH479179 EDW24286.1 UFQS01002223 UFQT01002223 SSX13534.1 SSX32963.1 CM000070 EAL28061.3 PSN49210.1 OUUW01000005 SPP80205.1 CH961022 KRF97397.1 GDHF01030205 GDHF01008287 JAI22109.1 JAI44027.1 APGK01029591 KB740735 ENN79223.1 CH964272 EDW84607.1 SPP80204.1 DS235004 EEB10247.1 CCAG010020230 GAKP01006417 JAC52535.1 JXJN01007734 KK852623 KDR19952.1 GBXI01008076 JAD06216.1 KB631809 ERL86314.1 KQ971372 EFA10554.2 NEVH01009379 PNF33299.1 GGMS01015665 MBY84868.1 GGMR01019675 MBY32294.1 AK402265 BAM18887.1 ABLF02039448 KPJ20268.1 AK401055 BAM17677.1 KPJ03632.1
KPJ03629.1 KZ149905 PZC78354.1 ODYU01000169 SOQ34484.1 AGBW02012152 OWR45561.1 DS231991 EDS30690.1 ATLV01014432 KE524972 KFB38742.1 JXUM01054123 KQ561806 KXJ77475.1 CH477298 EAT44319.1 ADMH02000786 ETN65057.1 AXCM01010351 JXUM01059338 JXUM01059339 PYGN01000289 PSN49208.1 AAAB01008880 EDO64173.2 APCN01002561 AXCN02001159 KK854089 PTY11054.1 CVRI01000003 CRK87120.1 ACPB03007640 CH940656 EDW58520.1 CH902617 EDV43720.1 CM000364 EDX11845.1 CH954181 EDV48006.1 CM000160 EDW97840.1 AJVK01013609 CH480821 EDW55074.1 JRES01001623 KNC21375.1 CH916377 EDV90713.1 AE014297 BT100070 AAF51898.1 ACX61611.1 CH933806 EDW14702.1 CP012526 ALC46825.1 AJWK01005739 CH479179 EDW24286.1 UFQS01002223 UFQT01002223 SSX13534.1 SSX32963.1 CM000070 EAL28061.3 PSN49210.1 OUUW01000005 SPP80205.1 CH961022 KRF97397.1 GDHF01030205 GDHF01008287 JAI22109.1 JAI44027.1 APGK01029591 KB740735 ENN79223.1 CH964272 EDW84607.1 SPP80204.1 DS235004 EEB10247.1 CCAG010020230 GAKP01006417 JAC52535.1 JXJN01007734 KK852623 KDR19952.1 GBXI01008076 JAD06216.1 KB631809 ERL86314.1 KQ971372 EFA10554.2 NEVH01009379 PNF33299.1 GGMS01015665 MBY84868.1 GGMR01019675 MBY32294.1 AK402265 BAM18887.1 ABLF02039448 KPJ20268.1 AK401055 BAM17677.1 KPJ03632.1
Proteomes
UP000005204
UP000053240
UP000037510
UP000053268
UP000007151
UP000002320
+ More
UP000030765 UP000069940 UP000249989 UP000008820 UP000075880 UP000069272 UP000000673 UP000075920 UP000075883 UP000076408 UP000245037 UP000075885 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075900 UP000075884 UP000075886 UP000183832 UP000015103 UP000008792 UP000192221 UP000007801 UP000000304 UP000008711 UP000002282 UP000092462 UP000001292 UP000037069 UP000001070 UP000000803 UP000009192 UP000092553 UP000092461 UP000008744 UP000095300 UP000001819 UP000268350 UP000007798 UP000019118 UP000009046 UP000092444 UP000092445 UP000095301 UP000092460 UP000092443 UP000078200 UP000027135 UP000030742 UP000007266 UP000235965 UP000091820 UP000007819
UP000030765 UP000069940 UP000249989 UP000008820 UP000075880 UP000069272 UP000000673 UP000075920 UP000075883 UP000076408 UP000245037 UP000075885 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075900 UP000075884 UP000075886 UP000183832 UP000015103 UP000008792 UP000192221 UP000007801 UP000000304 UP000008711 UP000002282 UP000092462 UP000001292 UP000037069 UP000001070 UP000000803 UP000009192 UP000092553 UP000092461 UP000008744 UP000095300 UP000001819 UP000268350 UP000007798 UP000019118 UP000009046 UP000092444 UP000092445 UP000095301 UP000092460 UP000092443 UP000078200 UP000027135 UP000030742 UP000007266 UP000235965 UP000091820 UP000007819
Pfam
PF07898 DUF1676
Interpro
IPR012464
DUF1676
ProteinModelPortal
H9IRY8
A0A0N1IH62
A0A0L7KXE4
A0A194QDY0
A0A2W1BXJ0
A0A2H1V2I8
+ More
A0A212EVS1 B0WLV5 A0A084VL98 A0A182GBT1 A0A1S4F7C9 Q17DB0 A0A182IN33 A0A182F6Q2 W5JL05 A0A182VSC9 A0A182MTD6 A0A182GF06 A0A182YGJ4 A0A2P8YY80 A0A182PF66 A0A182VAW6 A0A182THY0 A0A182XIZ6 A0A182L7G8 A7USY6 A0A182I7W1 A0A182RYJ9 A0A182N5P5 A0A182QTI9 A0A2R7VU74 A0A1J1HGH6 T1HNE3 B4MBP9 A0A1W4URM9 B3LVU6 B4QY88 B3NYM5 B4PVN1 A0A1B0DBB2 B4I4D1 A0A0L0BMR2 B4JYF1 Q9VNN1 B4KBM5 A0A0M4F4T2 A0A1B0GHW4 B4G3A4 A0A1I8PD50 A0A336L7Z4 Q297Z5 A0A2P8YY81 A0A3B0JDQ7 A0A0Q9WNG2 A0A0K8U631 N6TMG8 B4NHD1 A0A3B0JCU6 E0VA41 A0A1B0FJ43 A0A1A9ZSX7 A0A1I8MJ29 A0A034WEG0 A0A1B0B2Y0 A0A1A9YQ75 A0A1A9VDN7 A0A067R8N4 A0A0A1X5R7 U4TXI5 D6X0T8 A0A2J7QXJ5 A0A2S2R4H0 A0A1A9W5N3 A0A2S2PSH4 I4DLU7 J9JS56 A0A0N1PJF6 I4DID7
A0A212EVS1 B0WLV5 A0A084VL98 A0A182GBT1 A0A1S4F7C9 Q17DB0 A0A182IN33 A0A182F6Q2 W5JL05 A0A182VSC9 A0A182MTD6 A0A182GF06 A0A182YGJ4 A0A2P8YY80 A0A182PF66 A0A182VAW6 A0A182THY0 A0A182XIZ6 A0A182L7G8 A7USY6 A0A182I7W1 A0A182RYJ9 A0A182N5P5 A0A182QTI9 A0A2R7VU74 A0A1J1HGH6 T1HNE3 B4MBP9 A0A1W4URM9 B3LVU6 B4QY88 B3NYM5 B4PVN1 A0A1B0DBB2 B4I4D1 A0A0L0BMR2 B4JYF1 Q9VNN1 B4KBM5 A0A0M4F4T2 A0A1B0GHW4 B4G3A4 A0A1I8PD50 A0A336L7Z4 Q297Z5 A0A2P8YY81 A0A3B0JDQ7 A0A0Q9WNG2 A0A0K8U631 N6TMG8 B4NHD1 A0A3B0JCU6 E0VA41 A0A1B0FJ43 A0A1A9ZSX7 A0A1I8MJ29 A0A034WEG0 A0A1B0B2Y0 A0A1A9YQ75 A0A1A9VDN7 A0A067R8N4 A0A0A1X5R7 U4TXI5 D6X0T8 A0A2J7QXJ5 A0A2S2R4H0 A0A1A9W5N3 A0A2S2PSH4 I4DLU7 J9JS56 A0A0N1PJF6 I4DID7
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.989391
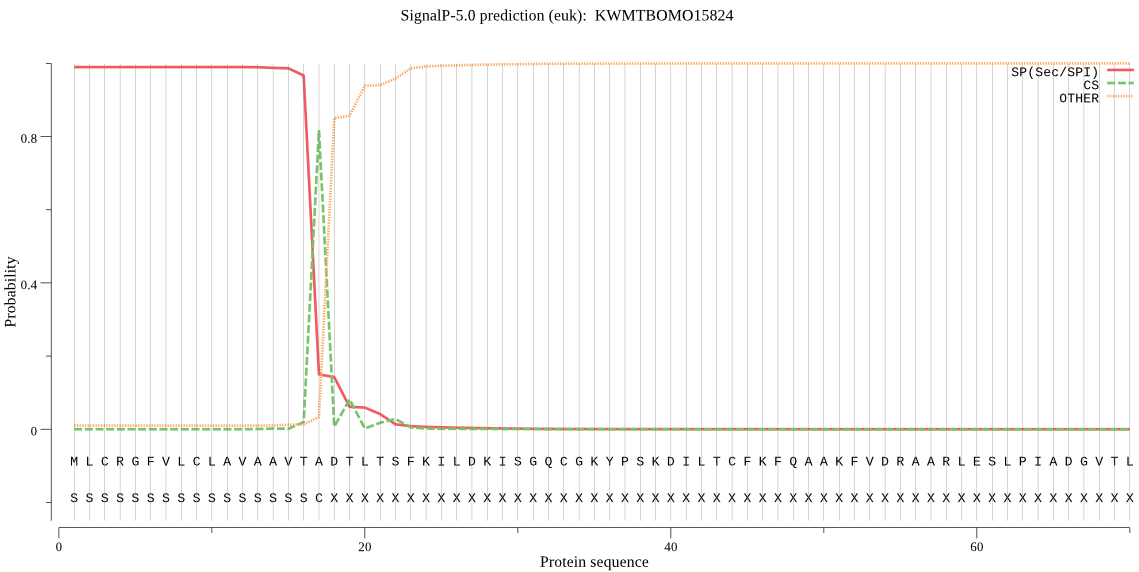
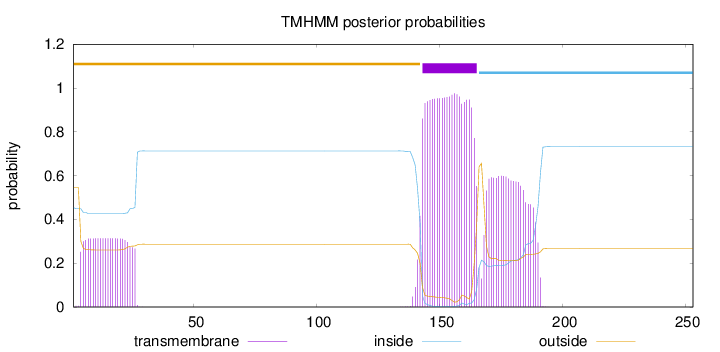
Length:
253
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
41.52145
Exp number, first 60 AAs:
6.98432
Total prob of N-in:
0.45372
outside
1 - 142
TMhelix
143 - 165
inside
166 - 253
Population Genetic Test Statistics
Pi
154.517673
Theta
135.696542
Tajima's D
1.179285
CLR
0.008198
CSRT
0.708764561771911
Interpretation
Uncertain
