Pre Gene Modal
BGIBMGA000012
Annotation
Osiris_9C_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 1.275
Sequence
CDS
ATGCTGAAGTACATCGCGTTATTAGCCTTGACGGCGTCAGTGCAGTGCAACCCGCTGAAAGAGAACAGCATATCGGAAAATTTAGTGGGAGTTATATCGGAGTGCATCGAAAGAGATACATCTTTGTGTATTAAGGAAAAAGCTCTTAAGTTTACCGAACGACTGGCCTTTTCTAAGGACATGAATATCTTCGATGGCATGTCTCTGGTCAACATTGGATCGGCCCGATCTGCCCGCAGCTACGAGCCTTTGGCCGAGGACCCAAAAGCTCGGGAATTGCAACTAGACGAGAGGATAGCCGACAATATGGGTGATTTCTTGGAGAACCATGTTATTCAACTTCGTTTGTCTGAGCCTGAAGCCGAGTCTAGGAGTTTGGATGATGAAGCTCGTGGCAAGAAAAAGAAGAAGCTGAAGCAGCTCCTTCCTCTTCTTCTCCTTTTGAAGTTGAAGCTGGCAGCTCTGATCCCACTTTTCCTGGGTATCATTGCCTTTGTTGCTGTTAAAGCAGTGTTCCTTGGCAAGATTGCCTTTGCAATGAGCGCTTTCCAACTCATCAGAAAACTTCTTAACAAGAACCAGTCTTCGGGAAGTTCCAGTATCTCATACGCTGCTCCTCATCACCACGAGGAGCATCCAGGATACTCCTACGAACCTGCCAGCAGCGGAGGCTGGGGTAGACAAGCCACTGACGCCCAATCATTGGCGTACGCCGGTCAATTGAACCACTAA
Protein
MLKYIALLALTASVQCNPLKENSISENLVGVISECIERDTSLCIKEKALKFTERLAFSKDMNIFDGMSLVNIGSARSARSYEPLAEDPKARELQLDERIADNMGDFLENHVIQLRLSEPEAESRSLDDEARGKKKKKLKQLLPLLLLLKLKLAALIPLFLGIIAFVAVKAVFLGKIAFAMSAFQLIRKLLNKNQSSGSSSISYAAPHHHEEHPGYSYEPASSGGWGRQATDAQSLAYAGQLNH
Summary
Uniprot
H9IRZ2
A0A2W1BTI8
A0A2A4J4F3
A0A0L7L0U4
A0A2H1V0Z6
A0A3S2NT33
+ More
A0A194QDY5 A0A212FGL5 A0A0N1IB67 I4DKL5 I4DIG3 A0A212FGK0 A0A0N0PEP2 A0A2H1V0W5 I4DM44 I4DID7 A0A0N1PJF6 I4DLU7 H9IRZ1 A0A212FGK5 A0A3S2TEL3 A0A2W1BZR2 A0A0L7L1B9 A0A2W1BVI2 A0A0L7L1S3 I4DLX5 A0A2H1V0X5 A0A212FC79 A0A2H1WVC9 B6DXB0 A0A2W1C052 A0A3S2L1Z5 I4DIF7 A0A0N1IQ79 A0A0N1IQF4 I4DIJ0 I4DLV7 H9IRZ3 A0A0L7L0S9 A0A2S2QFW6 A0A0C9R515 A0A232F323 K7IU81 A0A195DJ34 E2API8 A0A0K8VVA2 A0A0A1WIS1 A0A034WCK0 A0A1B0DBB0 A0A195FIY7 A0A195C1D8 A0A3S2NCE7 F4X739 E9IVC9 A0A0J7KXP8 E2B2W7 A0A0L7KRW0 Q17DA8 B0WLV8 A0A310SJB0 A0A151XHK0 J9JW80 W8BG86 H9IS19 A0A1J1HI88 A0A195B1E3 A0A158NDA7 A0A0L0BMW8 A0A026WHT7 A0A3B0K9I6 B4JYF3 A0A212FC98 A0A1I8ND72 A0A336MWX4 A0A1A9ZSW6 A0A1A9VIW5 B4MBQ2 A0A1A9YQ59 A0A1B0B2X3 A0A182F6Q0 A0A0Q9WVC3 A0A0M4EFA0 B4G3A2 Q297Z3 A0A1I8NZE3 B4KBM3 A0A2S2NVH5 A0A1A9W5M7 E0VA37 A0A2W1BYN3 A0A1Y9INF0 A0A182VGK2 A0A182L7H0 A0A182XIZ3
A0A194QDY5 A0A212FGL5 A0A0N1IB67 I4DKL5 I4DIG3 A0A212FGK0 A0A0N0PEP2 A0A2H1V0W5 I4DM44 I4DID7 A0A0N1PJF6 I4DLU7 H9IRZ1 A0A212FGK5 A0A3S2TEL3 A0A2W1BZR2 A0A0L7L1B9 A0A2W1BVI2 A0A0L7L1S3 I4DLX5 A0A2H1V0X5 A0A212FC79 A0A2H1WVC9 B6DXB0 A0A2W1C052 A0A3S2L1Z5 I4DIF7 A0A0N1IQ79 A0A0N1IQF4 I4DIJ0 I4DLV7 H9IRZ3 A0A0L7L0S9 A0A2S2QFW6 A0A0C9R515 A0A232F323 K7IU81 A0A195DJ34 E2API8 A0A0K8VVA2 A0A0A1WIS1 A0A034WCK0 A0A1B0DBB0 A0A195FIY7 A0A195C1D8 A0A3S2NCE7 F4X739 E9IVC9 A0A0J7KXP8 E2B2W7 A0A0L7KRW0 Q17DA8 B0WLV8 A0A310SJB0 A0A151XHK0 J9JW80 W8BG86 H9IS19 A0A1J1HI88 A0A195B1E3 A0A158NDA7 A0A0L0BMW8 A0A026WHT7 A0A3B0K9I6 B4JYF3 A0A212FC98 A0A1I8ND72 A0A336MWX4 A0A1A9ZSW6 A0A1A9VIW5 B4MBQ2 A0A1A9YQ59 A0A1B0B2X3 A0A182F6Q0 A0A0Q9WVC3 A0A0M4EFA0 B4G3A2 Q297Z3 A0A1I8NZE3 B4KBM3 A0A2S2NVH5 A0A1A9W5M7 E0VA37 A0A2W1BYN3 A0A1Y9INF0 A0A182VGK2 A0A182L7H0 A0A182XIZ3
Pubmed
EMBL
BABH01035639
KZ149905
PZC78349.1
NWSH01003095
PCG66955.1
JTDY01003716
+ More
KOB69102.1 ODYU01000169 SOQ34491.1 RSAL01000230 RVE43859.1 KQ459144 KPJ03634.1 AGBW02008652 OWR52867.1 KQ459717 KPJ20270.1 AK401833 BAM18455.1 AK401081 BAM17703.1 KPJ03633.1 OWR52866.1 KPJ20269.1 SOQ34490.1 AK402362 BAM18984.1 AK401055 BAM17677.1 KPJ03632.1 KPJ20268.1 AK402265 BAM18887.1 OWR52865.1 RVE43860.1 PZC78350.1 JTDY01003657 KOB69205.1 PZC78351.1 KOB69204.1 AK402293 BAM18915.1 SOQ34488.1 AGBW02009210 OWR51344.1 ODYU01011341 SOQ57029.1 FJ176298 ACI23620.1 PZC78346.1 RVE43856.1 AK401075 BAM17697.1 KPJ03637.1 KPJ20271.1 KPJ20273.1 AK401108 BAM17730.1 KPJ03635.1 AK402275 BAM18897.1 BABH01035638 KOB69103.1 GGMS01007442 MBY76645.1 GBYB01011404 JAG81171.1 NNAY01001166 OXU24919.1 AAZX01002662 KQ980800 KYN12856.1 GL441542 EFN64644.1 GDHF01009516 JAI42798.1 GBXI01015882 JAC98409.1 GAKP01007097 JAC51855.1 AJVK01013607 KQ981560 KYN39949.1 KQ978379 KYM94430.1 RVE43858.1 GL888828 EGI57651.1 GL766194 EFZ15457.1 LBMM01002063 KMQ95322.1 GL445250 EFN89936.1 JTDY01006624 KOB65825.1 CH477298 EAT44321.1 DS231991 EDS30693.1 KQ759821 OAD62711.1 KQ982130 KYQ59795.1 ABLF02039450 GAMC01010617 JAB95938.1 CVRI01000003 CRK87124.1 KQ976690 KYM78019.1 ADTU01012537 JRES01001623 KNC21445.1 KK107199 QOIP01000009 EZA55590.1 RLU18751.1 OUUW01000005 SPP80208.1 CH916377 EDV90715.1 OWR51343.1 UFQT01002904 SSX34245.1 CH940656 EDW58523.1 JXJN01007734 CH964272 KRG00079.1 CP012526 ALC46828.1 CH479179 EDW24284.1 CM000070 EAL28060.1 CH933806 EDW14700.1 GGMR01008571 MBY21190.1 DS235004 EEB10243.1 PZC78347.1
KOB69102.1 ODYU01000169 SOQ34491.1 RSAL01000230 RVE43859.1 KQ459144 KPJ03634.1 AGBW02008652 OWR52867.1 KQ459717 KPJ20270.1 AK401833 BAM18455.1 AK401081 BAM17703.1 KPJ03633.1 OWR52866.1 KPJ20269.1 SOQ34490.1 AK402362 BAM18984.1 AK401055 BAM17677.1 KPJ03632.1 KPJ20268.1 AK402265 BAM18887.1 OWR52865.1 RVE43860.1 PZC78350.1 JTDY01003657 KOB69205.1 PZC78351.1 KOB69204.1 AK402293 BAM18915.1 SOQ34488.1 AGBW02009210 OWR51344.1 ODYU01011341 SOQ57029.1 FJ176298 ACI23620.1 PZC78346.1 RVE43856.1 AK401075 BAM17697.1 KPJ03637.1 KPJ20271.1 KPJ20273.1 AK401108 BAM17730.1 KPJ03635.1 AK402275 BAM18897.1 BABH01035638 KOB69103.1 GGMS01007442 MBY76645.1 GBYB01011404 JAG81171.1 NNAY01001166 OXU24919.1 AAZX01002662 KQ980800 KYN12856.1 GL441542 EFN64644.1 GDHF01009516 JAI42798.1 GBXI01015882 JAC98409.1 GAKP01007097 JAC51855.1 AJVK01013607 KQ981560 KYN39949.1 KQ978379 KYM94430.1 RVE43858.1 GL888828 EGI57651.1 GL766194 EFZ15457.1 LBMM01002063 KMQ95322.1 GL445250 EFN89936.1 JTDY01006624 KOB65825.1 CH477298 EAT44321.1 DS231991 EDS30693.1 KQ759821 OAD62711.1 KQ982130 KYQ59795.1 ABLF02039450 GAMC01010617 JAB95938.1 CVRI01000003 CRK87124.1 KQ976690 KYM78019.1 ADTU01012537 JRES01001623 KNC21445.1 KK107199 QOIP01000009 EZA55590.1 RLU18751.1 OUUW01000005 SPP80208.1 CH916377 EDV90715.1 OWR51343.1 UFQT01002904 SSX34245.1 CH940656 EDW58523.1 JXJN01007734 CH964272 KRG00079.1 CP012526 ALC46828.1 CH479179 EDW24284.1 CM000070 EAL28060.1 CH933806 EDW14700.1 GGMR01008571 MBY21190.1 DS235004 EEB10243.1 PZC78347.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000283053
UP000053268
UP000007151
+ More
UP000053240 UP000215335 UP000002358 UP000078492 UP000000311 UP000092462 UP000078541 UP000078542 UP000007755 UP000036403 UP000008237 UP000008820 UP000002320 UP000075809 UP000007819 UP000183832 UP000078540 UP000005205 UP000037069 UP000053097 UP000279307 UP000268350 UP000001070 UP000095301 UP000092445 UP000078200 UP000008792 UP000092443 UP000092460 UP000069272 UP000007798 UP000092553 UP000008744 UP000001819 UP000095300 UP000009192 UP000091820 UP000009046 UP000075902 UP000075903 UP000075882 UP000076407
UP000053240 UP000215335 UP000002358 UP000078492 UP000000311 UP000092462 UP000078541 UP000078542 UP000007755 UP000036403 UP000008237 UP000008820 UP000002320 UP000075809 UP000007819 UP000183832 UP000078540 UP000005205 UP000037069 UP000053097 UP000279307 UP000268350 UP000001070 UP000095301 UP000092445 UP000078200 UP000008792 UP000092443 UP000092460 UP000069272 UP000007798 UP000092553 UP000008744 UP000001819 UP000095300 UP000009192 UP000091820 UP000009046 UP000075902 UP000075903 UP000075882 UP000076407
Pfam
PF07898 DUF1676
ProteinModelPortal
H9IRZ2
A0A2W1BTI8
A0A2A4J4F3
A0A0L7L0U4
A0A2H1V0Z6
A0A3S2NT33
+ More
A0A194QDY5 A0A212FGL5 A0A0N1IB67 I4DKL5 I4DIG3 A0A212FGK0 A0A0N0PEP2 A0A2H1V0W5 I4DM44 I4DID7 A0A0N1PJF6 I4DLU7 H9IRZ1 A0A212FGK5 A0A3S2TEL3 A0A2W1BZR2 A0A0L7L1B9 A0A2W1BVI2 A0A0L7L1S3 I4DLX5 A0A2H1V0X5 A0A212FC79 A0A2H1WVC9 B6DXB0 A0A2W1C052 A0A3S2L1Z5 I4DIF7 A0A0N1IQ79 A0A0N1IQF4 I4DIJ0 I4DLV7 H9IRZ3 A0A0L7L0S9 A0A2S2QFW6 A0A0C9R515 A0A232F323 K7IU81 A0A195DJ34 E2API8 A0A0K8VVA2 A0A0A1WIS1 A0A034WCK0 A0A1B0DBB0 A0A195FIY7 A0A195C1D8 A0A3S2NCE7 F4X739 E9IVC9 A0A0J7KXP8 E2B2W7 A0A0L7KRW0 Q17DA8 B0WLV8 A0A310SJB0 A0A151XHK0 J9JW80 W8BG86 H9IS19 A0A1J1HI88 A0A195B1E3 A0A158NDA7 A0A0L0BMW8 A0A026WHT7 A0A3B0K9I6 B4JYF3 A0A212FC98 A0A1I8ND72 A0A336MWX4 A0A1A9ZSW6 A0A1A9VIW5 B4MBQ2 A0A1A9YQ59 A0A1B0B2X3 A0A182F6Q0 A0A0Q9WVC3 A0A0M4EFA0 B4G3A2 Q297Z3 A0A1I8NZE3 B4KBM3 A0A2S2NVH5 A0A1A9W5M7 E0VA37 A0A2W1BYN3 A0A1Y9INF0 A0A182VGK2 A0A182L7H0 A0A182XIZ3
A0A194QDY5 A0A212FGL5 A0A0N1IB67 I4DKL5 I4DIG3 A0A212FGK0 A0A0N0PEP2 A0A2H1V0W5 I4DM44 I4DID7 A0A0N1PJF6 I4DLU7 H9IRZ1 A0A212FGK5 A0A3S2TEL3 A0A2W1BZR2 A0A0L7L1B9 A0A2W1BVI2 A0A0L7L1S3 I4DLX5 A0A2H1V0X5 A0A212FC79 A0A2H1WVC9 B6DXB0 A0A2W1C052 A0A3S2L1Z5 I4DIF7 A0A0N1IQ79 A0A0N1IQF4 I4DIJ0 I4DLV7 H9IRZ3 A0A0L7L0S9 A0A2S2QFW6 A0A0C9R515 A0A232F323 K7IU81 A0A195DJ34 E2API8 A0A0K8VVA2 A0A0A1WIS1 A0A034WCK0 A0A1B0DBB0 A0A195FIY7 A0A195C1D8 A0A3S2NCE7 F4X739 E9IVC9 A0A0J7KXP8 E2B2W7 A0A0L7KRW0 Q17DA8 B0WLV8 A0A310SJB0 A0A151XHK0 J9JW80 W8BG86 H9IS19 A0A1J1HI88 A0A195B1E3 A0A158NDA7 A0A0L0BMW8 A0A026WHT7 A0A3B0K9I6 B4JYF3 A0A212FC98 A0A1I8ND72 A0A336MWX4 A0A1A9ZSW6 A0A1A9VIW5 B4MBQ2 A0A1A9YQ59 A0A1B0B2X3 A0A182F6Q0 A0A0Q9WVC3 A0A0M4EFA0 B4G3A2 Q297Z3 A0A1I8NZE3 B4KBM3 A0A2S2NVH5 A0A1A9W5M7 E0VA37 A0A2W1BYN3 A0A1Y9INF0 A0A182VGK2 A0A182L7H0 A0A182XIZ3
Ontologies
GO
PANTHER
Topology
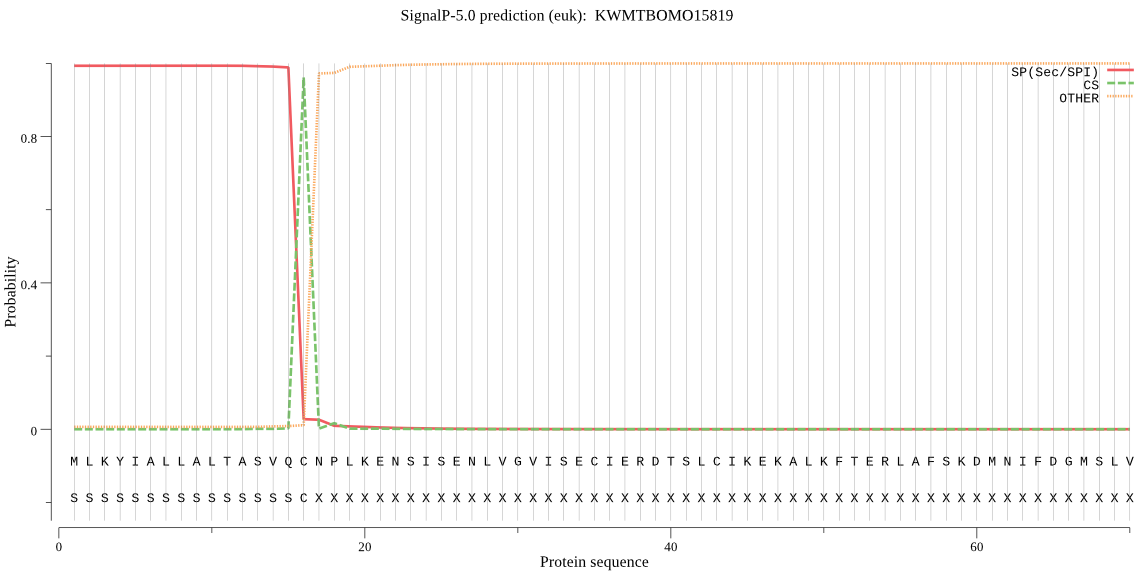
SignalP
Position: 1 - 16,
Likelihood: 0.992868
Length:
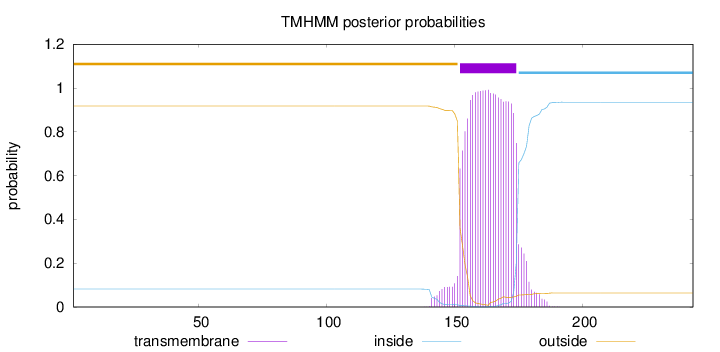
243
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.47025
Exp number, first 60 AAs:
0.00044
Total prob of N-in:
0.08169
outside
1 - 151
TMhelix
152 - 174
inside
175 - 243
Population Genetic Test Statistics
Pi
134.1129
Theta
119.227763
Tajima's D
-0.242821
CLR
0.01125
CSRT
0.297885105744713
Interpretation
Uncertain
