Gene
KWMTBOMO15815
Pre Gene Modal
BGIBMGA000044
Annotation
osiris_7_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.018 Nuclear Reliability : 1.127
Sequence
CDS
ATGTGGAGGACGTCGTTTATCTTCGCTTTGATTGCGGCTACATCCGCTCTGCCCGCCCAAGATACGTCGTACTCTGAAAACGATCTCGACGACTGCTTAAAGAAAGACCCTACATCCTGTCTGAAAAACAAGTTCTTCTCATACGTGGACAAGTATATAGGGCAGAAGGATTCGTTCGCTCTTACCGATGGAGTGAAGATCTACAAGTCAGCTGATGTCATACAGACGGGAGCACCCAGATCTCTCGACCCACTTTCAAGACTGAAGTCCTATTTGGAAACTCATTCTCTTAGTGTTGAAGTGAAAGGTACTGATATTTTGGATGCTGTGACTGGTGCAGGAAGAGCTCTTCAAGATACTGTGACTTCCTTCGTCGATGATGACGCCGCGGAAGAAAGCAGAGGGAAAAAGAAGAAAGCCTCAAAAATCCTTGGACCAATAATGGCGATTATGGCTATGAAGATGATGGCAATAATGCCTCTTGCGATCGGAGCTATAGCATTAATAGCTGGCAAGGCGTTACTGATAGGAAAACTGGCATTGGTCTTGTCAGCGATAATTGGTTTGAAGAAATTGATGTCACAACAGAAGCACGTGACTTATGAAGTGGTGGCGCATCCTCAGCATACATCTAGTCACACTTCGAGCCACGATTATGGAGGTACAAGCGGATATGGCGGAGACTCGTCTAGTGGTTACAGTAGCGGCGGAGGTCACGGAAGCAGCGGATGGGGTAGGTCATTAGACGCGCAGAGCCTAGCGTATTCAGCCCACAAACCCCATTAG
Protein
MWRTSFIFALIAATSALPAQDTSYSENDLDDCLKKDPTSCLKNKFFSYVDKYIGQKDSFALTDGVKIYKSADVIQTGAPRSLDPLSRLKSYLETHSLSVEVKGTDILDAVTGAGRALQDTVTSFVDDDAAEESRGKKKKASKILGPIMAIMAMKMMAIMPLAIGAIALIAGKALLIGKLALVLSAIIGLKKLMSQQKHVTYEVVAHPQHTSSHTSSHDYGGTSGYGGDSSSGYSSGGGHGSSGWGRSLDAQSLAYSAHKPH
Summary
Uniprot
B6VGH6
A0A2H1V201
A0A212FC82
A0A0N1IK14
A0A194QDL4
A0A0L7L9C6
+ More
I4DN95 A0A2W1BXH4 I4DR98 A0A1W4XN02 B4PVM7 B3NYM1 A0A1W4URS9 A0A0Y0DHV2 A0A109QMH5 A0A109QNS1 Q9VNM7 B3LVV0 A0A1B0DBA8 Q5U185 A0A1J1I169 A0A1B0GK54 A0A1I8P1N2 B4NHD4 A0A1B0CG45 B4KB20 B4MBQ5 A0A0M4EUM7 B4JYF5 A0A034VW33 A0A0K8TZH3 F4X741 A0A182VBD5 A0A182TUL1 A0A182L7H3 A0A182XIZ1 Q7QB91 A0A182I7W6 A0A0L0BN52 Q297Z1 B4G3A0 A0A0A1XGR8 A0A1A9ZSZ0 A0A1A9VIW2 A0A1B0FET7 W8B803 D6X0U3 A0A3B0JXH7 A0A182RYJ4 A0A195FI64 A0A1A9YQ72 A0A1B0B2V9 A0A182JJ26 A0A182MQX5 A0A182GQU4 A0A182YGI9 A0A1I8NCR2 A0A182VSC4 A0A182Q8N1 A0A182PF61 A0A1A9W5M4 A0A182N5P0 A0A154PH04 A0A182JU31 A0A0N0U7Q3 A0A0C9PY25 A0A088AGF6 A0A310SV62 B4I4C7 A0A182TCJ4 A0A0L7RJD4 B0WLW0 A0A195DIZ3 A0A151XHB5 A0A195B152 A0A158NDA6 A0A195C0R0 A0A0J7KX06 W5JAM2 A0A084VL87 A0A026WHW0 A0A2A3EQU9 A0A3L8DFS3 E2B2W6 A0A182F6P8 E0VA36 J3JVW2 A0A336N0N1 K7IU79 E2API9 A0A2P8YFV4 A0A1Y1LKN4 A0A067R8W9 T1HNC7 A0A170Z9Q0
I4DN95 A0A2W1BXH4 I4DR98 A0A1W4XN02 B4PVM7 B3NYM1 A0A1W4URS9 A0A0Y0DHV2 A0A109QMH5 A0A109QNS1 Q9VNM7 B3LVV0 A0A1B0DBA8 Q5U185 A0A1J1I169 A0A1B0GK54 A0A1I8P1N2 B4NHD4 A0A1B0CG45 B4KB20 B4MBQ5 A0A0M4EUM7 B4JYF5 A0A034VW33 A0A0K8TZH3 F4X741 A0A182VBD5 A0A182TUL1 A0A182L7H3 A0A182XIZ1 Q7QB91 A0A182I7W6 A0A0L0BN52 Q297Z1 B4G3A0 A0A0A1XGR8 A0A1A9ZSZ0 A0A1A9VIW2 A0A1B0FET7 W8B803 D6X0U3 A0A3B0JXH7 A0A182RYJ4 A0A195FI64 A0A1A9YQ72 A0A1B0B2V9 A0A182JJ26 A0A182MQX5 A0A182GQU4 A0A182YGI9 A0A1I8NCR2 A0A182VSC4 A0A182Q8N1 A0A182PF61 A0A1A9W5M4 A0A182N5P0 A0A154PH04 A0A182JU31 A0A0N0U7Q3 A0A0C9PY25 A0A088AGF6 A0A310SV62 B4I4C7 A0A182TCJ4 A0A0L7RJD4 B0WLW0 A0A195DIZ3 A0A151XHB5 A0A195B152 A0A158NDA6 A0A195C0R0 A0A0J7KX06 W5JAM2 A0A084VL87 A0A026WHW0 A0A2A3EQU9 A0A3L8DFS3 E2B2W6 A0A182F6P8 E0VA36 J3JVW2 A0A336N0N1 K7IU79 E2API9 A0A2P8YFV4 A0A1Y1LKN4 A0A067R8W9 T1HNC7 A0A170Z9Q0
Pubmed
19121390
22118469
26354079
26227816
22651552
28756777
+ More
17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 21719571 20966253 12364791 14747013 17210077 26108605 15632085 25830018 24495485 18362917 19820115 26483478 25244985 25315136 21347285 20920257 23761445 24438588 24508170 30249741 20798317 20566863 22516182 23537049 20075255 29403074 28004739 24845553
17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 21719571 20966253 12364791 14747013 17210077 26108605 15632085 25830018 24495485 18362917 19820115 26483478 25244985 25315136 21347285 20920257 23761445 24438588 24508170 30249741 20798317 20566863 22516182 23537049 20075255 29403074 28004739 24845553
EMBL
BABH01035637
FJ392544
ACJ12371.1
ODYU01000127
SOQ34382.1
AGBW02009210
+ More
OWR51345.1 KQ459717 KPJ20274.1 KQ459144 KPJ03638.1 JTDY01002231 KOB71846.1 AK402783 BAM19385.1 KZ149905 PZC78345.1 AK405030 BAM20438.1 CM000160 EDW97836.1 CH954181 EDV48002.1 KT388095 AMB21537.1 KT388097 AMB21539.1 KT388094 AMB21536.1 AE014297 BT056248 AAF51902.1 ACL68695.1 CH902617 EDV43724.1 AJVK01013604 BT016007 AAV36892.1 CVRI01000023 CRK92113.1 AJWK01022600 CH964272 EDW84610.1 AJWK01010740 CH933806 EDW14698.1 CH940656 EDW58526.1 CP012526 ALC46830.1 CH916377 EDV90717.1 GAKP01012640 JAC46312.1 GDHF01032641 JAI19673.1 GL888828 EGI57653.1 AAAB01008880 EAA08668.1 APCN01002562 JRES01001623 KNC21432.1 CM000070 EAL28064.2 CH479179 EDW24282.1 GBXI01017301 GBXI01004177 JAC96990.1 JAD10115.1 CCAG010019117 GAMC01013372 JAB93183.1 KQ971372 EFA09550.1 OUUW01000005 SPP80210.1 KQ981560 KYN39947.1 JXJN01007733 AXCM01005108 JXUM01015764 JXUM01067724 KQ562465 KQ560439 KXJ75850.1 KXJ82399.1 AXCN02001159 KQ434902 KZC11141.1 KQ435706 KOX80174.1 GBYB01006433 JAG76200.1 KQ759821 OAD62709.1 CH480821 EDW55070.1 KQ414581 KOC70944.1 DS231991 EDS30695.1 KQ980800 KYN12858.1 KQ982130 KYQ59793.1 KQ976690 KYM78022.1 ADTU01012535 KQ978379 KYM94432.1 LBMM01002459 KMQ94804.1 ADMH02001809 ETN61046.1 ATLV01014428 KE524972 KFB38731.1 KK107199 EZA55588.1 KZ288200 PBC33596.1 QOIP01000009 RLU18748.1 GL445250 EFN89935.1 DS235004 EEB10242.1 APGK01029587 BT127380 KB740735 KB631809 AEE62342.1 ENN79218.1 ERL86309.1 UFQT01002904 SSX34247.1 AAZX01002662 GL441542 EFN64645.1 PYGN01000628 PSN43137.1 GEZM01053043 JAV74212.1 KK852623 KDR19948.1 ACPB03007640 GEMB01002459 JAS00727.1
OWR51345.1 KQ459717 KPJ20274.1 KQ459144 KPJ03638.1 JTDY01002231 KOB71846.1 AK402783 BAM19385.1 KZ149905 PZC78345.1 AK405030 BAM20438.1 CM000160 EDW97836.1 CH954181 EDV48002.1 KT388095 AMB21537.1 KT388097 AMB21539.1 KT388094 AMB21536.1 AE014297 BT056248 AAF51902.1 ACL68695.1 CH902617 EDV43724.1 AJVK01013604 BT016007 AAV36892.1 CVRI01000023 CRK92113.1 AJWK01022600 CH964272 EDW84610.1 AJWK01010740 CH933806 EDW14698.1 CH940656 EDW58526.1 CP012526 ALC46830.1 CH916377 EDV90717.1 GAKP01012640 JAC46312.1 GDHF01032641 JAI19673.1 GL888828 EGI57653.1 AAAB01008880 EAA08668.1 APCN01002562 JRES01001623 KNC21432.1 CM000070 EAL28064.2 CH479179 EDW24282.1 GBXI01017301 GBXI01004177 JAC96990.1 JAD10115.1 CCAG010019117 GAMC01013372 JAB93183.1 KQ971372 EFA09550.1 OUUW01000005 SPP80210.1 KQ981560 KYN39947.1 JXJN01007733 AXCM01005108 JXUM01015764 JXUM01067724 KQ562465 KQ560439 KXJ75850.1 KXJ82399.1 AXCN02001159 KQ434902 KZC11141.1 KQ435706 KOX80174.1 GBYB01006433 JAG76200.1 KQ759821 OAD62709.1 CH480821 EDW55070.1 KQ414581 KOC70944.1 DS231991 EDS30695.1 KQ980800 KYN12858.1 KQ982130 KYQ59793.1 KQ976690 KYM78022.1 ADTU01012535 KQ978379 KYM94432.1 LBMM01002459 KMQ94804.1 ADMH02001809 ETN61046.1 ATLV01014428 KE524972 KFB38731.1 KK107199 EZA55588.1 KZ288200 PBC33596.1 QOIP01000009 RLU18748.1 GL445250 EFN89935.1 DS235004 EEB10242.1 APGK01029587 BT127380 KB740735 KB631809 AEE62342.1 ENN79218.1 ERL86309.1 UFQT01002904 SSX34247.1 AAZX01002662 GL441542 EFN64645.1 PYGN01000628 PSN43137.1 GEZM01053043 JAV74212.1 KK852623 KDR19948.1 ACPB03007640 GEMB01002459 JAS00727.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000037510
UP000192223
+ More
UP000002282 UP000008711 UP000192221 UP000000803 UP000007801 UP000092462 UP000183832 UP000092461 UP000095300 UP000007798 UP000009192 UP000008792 UP000092553 UP000001070 UP000007755 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000037069 UP000001819 UP000008744 UP000092445 UP000078200 UP000092444 UP000007266 UP000268350 UP000075900 UP000078541 UP000092443 UP000092460 UP000075880 UP000075883 UP000069940 UP000249989 UP000076408 UP000095301 UP000075920 UP000075886 UP000075885 UP000091820 UP000075884 UP000076502 UP000075881 UP000053105 UP000005203 UP000001292 UP000075901 UP000053825 UP000002320 UP000078492 UP000075809 UP000078540 UP000005205 UP000078542 UP000036403 UP000000673 UP000030765 UP000053097 UP000242457 UP000279307 UP000008237 UP000069272 UP000009046 UP000019118 UP000030742 UP000002358 UP000000311 UP000245037 UP000027135 UP000015103
UP000002282 UP000008711 UP000192221 UP000000803 UP000007801 UP000092462 UP000183832 UP000092461 UP000095300 UP000007798 UP000009192 UP000008792 UP000092553 UP000001070 UP000007755 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000037069 UP000001819 UP000008744 UP000092445 UP000078200 UP000092444 UP000007266 UP000268350 UP000075900 UP000078541 UP000092443 UP000092460 UP000075880 UP000075883 UP000069940 UP000249989 UP000076408 UP000095301 UP000075920 UP000075886 UP000075885 UP000091820 UP000075884 UP000076502 UP000075881 UP000053105 UP000005203 UP000001292 UP000075901 UP000053825 UP000002320 UP000078492 UP000075809 UP000078540 UP000005205 UP000078542 UP000036403 UP000000673 UP000030765 UP000053097 UP000242457 UP000279307 UP000008237 UP000069272 UP000009046 UP000019118 UP000030742 UP000002358 UP000000311 UP000245037 UP000027135 UP000015103
Pfam
PF07898 DUF1676
Interpro
IPR012464
DUF1676
ProteinModelPortal
B6VGH6
A0A2H1V201
A0A212FC82
A0A0N1IK14
A0A194QDL4
A0A0L7L9C6
+ More
I4DN95 A0A2W1BXH4 I4DR98 A0A1W4XN02 B4PVM7 B3NYM1 A0A1W4URS9 A0A0Y0DHV2 A0A109QMH5 A0A109QNS1 Q9VNM7 B3LVV0 A0A1B0DBA8 Q5U185 A0A1J1I169 A0A1B0GK54 A0A1I8P1N2 B4NHD4 A0A1B0CG45 B4KB20 B4MBQ5 A0A0M4EUM7 B4JYF5 A0A034VW33 A0A0K8TZH3 F4X741 A0A182VBD5 A0A182TUL1 A0A182L7H3 A0A182XIZ1 Q7QB91 A0A182I7W6 A0A0L0BN52 Q297Z1 B4G3A0 A0A0A1XGR8 A0A1A9ZSZ0 A0A1A9VIW2 A0A1B0FET7 W8B803 D6X0U3 A0A3B0JXH7 A0A182RYJ4 A0A195FI64 A0A1A9YQ72 A0A1B0B2V9 A0A182JJ26 A0A182MQX5 A0A182GQU4 A0A182YGI9 A0A1I8NCR2 A0A182VSC4 A0A182Q8N1 A0A182PF61 A0A1A9W5M4 A0A182N5P0 A0A154PH04 A0A182JU31 A0A0N0U7Q3 A0A0C9PY25 A0A088AGF6 A0A310SV62 B4I4C7 A0A182TCJ4 A0A0L7RJD4 B0WLW0 A0A195DIZ3 A0A151XHB5 A0A195B152 A0A158NDA6 A0A195C0R0 A0A0J7KX06 W5JAM2 A0A084VL87 A0A026WHW0 A0A2A3EQU9 A0A3L8DFS3 E2B2W6 A0A182F6P8 E0VA36 J3JVW2 A0A336N0N1 K7IU79 E2API9 A0A2P8YFV4 A0A1Y1LKN4 A0A067R8W9 T1HNC7 A0A170Z9Q0
I4DN95 A0A2W1BXH4 I4DR98 A0A1W4XN02 B4PVM7 B3NYM1 A0A1W4URS9 A0A0Y0DHV2 A0A109QMH5 A0A109QNS1 Q9VNM7 B3LVV0 A0A1B0DBA8 Q5U185 A0A1J1I169 A0A1B0GK54 A0A1I8P1N2 B4NHD4 A0A1B0CG45 B4KB20 B4MBQ5 A0A0M4EUM7 B4JYF5 A0A034VW33 A0A0K8TZH3 F4X741 A0A182VBD5 A0A182TUL1 A0A182L7H3 A0A182XIZ1 Q7QB91 A0A182I7W6 A0A0L0BN52 Q297Z1 B4G3A0 A0A0A1XGR8 A0A1A9ZSZ0 A0A1A9VIW2 A0A1B0FET7 W8B803 D6X0U3 A0A3B0JXH7 A0A182RYJ4 A0A195FI64 A0A1A9YQ72 A0A1B0B2V9 A0A182JJ26 A0A182MQX5 A0A182GQU4 A0A182YGI9 A0A1I8NCR2 A0A182VSC4 A0A182Q8N1 A0A182PF61 A0A1A9W5M4 A0A182N5P0 A0A154PH04 A0A182JU31 A0A0N0U7Q3 A0A0C9PY25 A0A088AGF6 A0A310SV62 B4I4C7 A0A182TCJ4 A0A0L7RJD4 B0WLW0 A0A195DIZ3 A0A151XHB5 A0A195B152 A0A158NDA6 A0A195C0R0 A0A0J7KX06 W5JAM2 A0A084VL87 A0A026WHW0 A0A2A3EQU9 A0A3L8DFS3 E2B2W6 A0A182F6P8 E0VA36 J3JVW2 A0A336N0N1 K7IU79 E2API9 A0A2P8YFV4 A0A1Y1LKN4 A0A067R8W9 T1HNC7 A0A170Z9Q0
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.997121
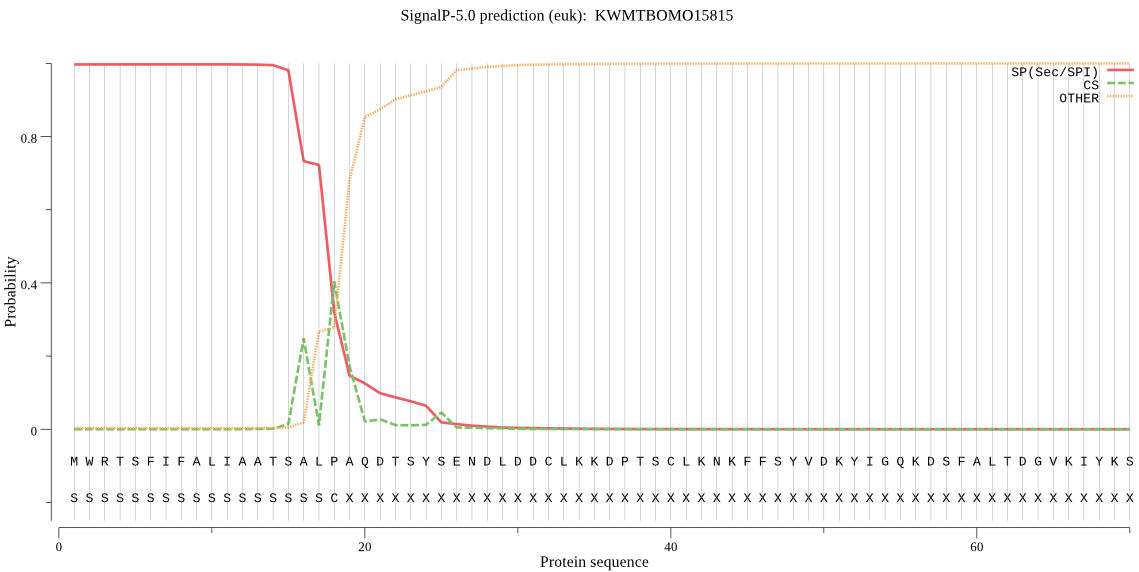
Length:
261
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
34.7934700000001
Exp number, first 60 AAs:
0.07517
Total prob of N-in:
0.57741
outside
1 - 160
TMhelix
161 - 183
inside
184 - 261

Population Genetic Test Statistics
Pi
316.245041
Theta
227.150945
Tajima's D
1.227423
CLR
0
CSRT
0.72336383180841
Interpretation
Uncertain
