Gene
KWMTBOMO15808
Pre Gene Modal
BGIBMGA000016
Annotation
neuropeptide_receptor_A4_[Bombyx_mori]
Full name
Neuropeptide F receptor
Location in the cell
PlasmaMembrane Reliability : 4.996
Sequence
CDS
ATGCTTACTGATGTTAGGGGGCTTGACCCCAGCGTGAACCTCACGTTGAATGCTTCAGCGAAACAGCTTATTGAGGCTTCAAAAGGCATCCAAGACCCAAACATACTCCTGGAGAAGTTTTCTCAGAATCGAAAAGTCGACGATCCAACGAGATCCTTACTGATAGCGTTTTACATCATCCTGGTCGTGATCGGAGCTGTGGGGAATGCACTCGTCATATTATCTGTGGTGCGAAAGCCGGTCATGAGAACAGCAAGGAACATGTTCATCGTCAATTTAGCCGTGTCCGATGCCCTGGTCTGCGTGGTCGGAACCCCGCTGACTCTTATGGAACTCCTGACCAAGCATTGGCCGTTACCGGACTGGCCAAGTTTGTGCAAGGCGTGTGGCGCGATCCAAGCTATATCGATCTTCGTGTCCACTATATCCATAACTGCCATTGCCCTTGATCGATATCAGTTGATCGTCTACCCCACCAAGCCTGGCGTTCAAACAATAGGAGCGCTTGTCACTATGTTCTTCATCTGGGTAACAGCATTCATCCTTGCTTCACCCCTCTACATCTTCCGGAGCCTCAAAACGCACAAACTCGGCATCGCCGGTATCAGCTCTCTAAGTTTCTGCATAGAAGACTGGCCTATCACTGACGGTAGAGCTATCTATTCTCTCCTAAGCCTGATCTTTCAATACCTTCTACCGGTTCTGGTCGTCGTGATGGCCCACATTCAGATACATAGAAGACTGCGAGGCCGAAGAAGGACCACGAGGAAAACGCCAGCAATATTAATAGCCATAGCAGTTACATACGTCATAAGCTGGCTACCACTTAATGTATTTAATCTGGTGGCTGACTTCAGTTCGGCGCCATTTAAAGACGAGAAGACTATGACGGTTACTTATGCTGTCTGCCACATGTTTGGCATGTCGAGTGCCGTTTCAAATCCTTTACTCTACGGATGGTTAAATGATAATTTCCGAAAGGAATTCGAAGAGATCCTGACGAAGTGCTGCTGCAGGAAGAAGCCTTTAGTTAACGGCACGAGGACTACTAATCGGAGGATGGAGACGGAATTGACTGCGCTGGCACAGTTGGAGCATACCGTGACCGGGAACACCAAGACGTCGCAGTGCTCCCAGGTTTTCTGA
Protein
MLTDVRGLDPSVNLTLNASAKQLIEASKGIQDPNILLEKFSQNRKVDDPTRSLLIAFYIILVVIGAVGNALVILSVVRKPVMRTARNMFIVNLAVSDALVCVVGTPLTLMELLTKHWPLPDWPSLCKACGAIQAISIFVSTISITAIALDRYQLIVYPTKPGVQTIGALVTMFFIWVTAFILASPLYIFRSLKTHKLGIAGISSLSFCIEDWPITDGRAIYSLLSLIFQYLLPVLVVVMAHIQIHRRLRGRRRTTRKTPAILIAIAVTYVISWLPLNVFNLVADFSSAPFKDEKTMTVTYAVCHMFGMSSAVSNPLLYGWLNDNFRKEFEEILTKCCCRKKPLVNGTRTTNRRMETELTALAQLEHTVTGNTKTSQCSQVF
Summary
Description
Receptor for NPF. Integral part of the sensory system that mediates food signaling, providing the neural basis for the regulation of food response; coordinates larval foraging and social behavior changes during development. Required in dopaminergic (DA) neurons that innervate the mushroom body for satiety to suppress appetitive memory performance; a key factor in the internal state of hunger in the brain. NPF neurons coordinately modulate diverse sensory and motor neurons important for feeding, flight, and locomotion. NPF/NPFR pathway exerts its suppressive effect on larval aversion to diverse stressful stimuli (chemical stress and noxious heat) through attenuation of TRP channel-induced neuronal excitation. NPF neural signaling system plays a physiological role in acute modulation of alcohol sensitivity in adults, rather than a general response to intoxication by sedative agents. Activation and inhibition of the NPF system reduces and enhances ethanol preference, respectively. Sexual experience, the NPF system activity and ethanol consumption are all linked; sexual deprivation is a major contributor to enhanced ethanol preference.
Similarity
Belongs to the G-protein coupled receptor 1 family.
Keywords
Alternative splicing
Behavior
Complete proteome
Disulfide bond
G-protein coupled receptor
Membrane
Neuropeptide
Receptor
Reference proteome
Stress response
Transducer
Transmembrane
Transmembrane helix
Feature
chain Neuropeptide F receptor
splice variant In isoform 2 and isoform 6.
splice variant In isoform 2 and isoform 6.
Uniprot
B3XXL6
A0A2A4J983
A0A0S1YD64
A0A2A4J9L8
A0A2W1BYM3
A0A194QDU4
+ More
A0A1E1W5R9 H9IRZ6 A0A212F140 A0A1E1WB26 A0A0N1IES3 A0A1W4XJH0 A0A2P8YDJ1 A0A067RKH6 A0A2J7QXK0 A0A1S4FQV5 B0WJ64 U5N0V1 A0A182H7A0 A0A1A9VIV6 A0A182GCN7 A0A1B0ERV5 Q16SC4 A0A3R7PJV0 B4G380 A0A1A9ZT02 A0A0R3NHE1 B4JYG2 Q297Y5 A0A0R3NMC1 A0A0R3NH99 A0A182XIY4 A0A182VIA4 A0A345AH67 A0A139WBY5 A0A182YGI2 A0A182L7I2 Q7PQN7 A0A182I7X3 A0A182RYI7 A0A182VSB6 A0A182P5Y9 A0A3B0JCV9 A0A182QAR7 A0A1I8NTF3 A0A182N5N3 A0A1A9W5L1 A0A182F6P1 B4MBR2 W5JC27 A0A1I8M4L5 A0A1I8M4K9 A0A1B0B4V9 A0A084VL74 A0A1A9YQ48 B4KB11 A0A1W4V450 A0A0R3NNU4 B4NHE1 E9GG50 A0A0M4F4U0 A0A0Q9VZP6 E9GBE9 A0A0P6CD76 A0A0Q9X5N6 A0A2P6LJY7 A0A0P6C9L9 A0A0P6HFJ0 T1IW42 A0A1D2N3W3 B3NYL4 E0VA32 A0A0P4Z0P0 W8C5G8 Q9VNM1-6 Q9VNM1 A0A0N7ZMC1 B4QXP8 Q9VNM1-4 A0A0B4K601 A0A0P5CAX0 A0A0P5YB18 A0A0P5WDL7 A0A0P5I5Y2 A0A0P5ZX39 A0A0P5MX59 A0A0N8DQ53 A0A164RXS6 A0A0P5MQL8 A0A0P6FUI5 A0A0P5WCS6 A0A0P5CZ61 A0A0R1E416 A0A0N8D8A5 B4PVM0 A0A0P4YK79 T1KR44
A0A1E1W5R9 H9IRZ6 A0A212F140 A0A1E1WB26 A0A0N1IES3 A0A1W4XJH0 A0A2P8YDJ1 A0A067RKH6 A0A2J7QXK0 A0A1S4FQV5 B0WJ64 U5N0V1 A0A182H7A0 A0A1A9VIV6 A0A182GCN7 A0A1B0ERV5 Q16SC4 A0A3R7PJV0 B4G380 A0A1A9ZT02 A0A0R3NHE1 B4JYG2 Q297Y5 A0A0R3NMC1 A0A0R3NH99 A0A182XIY4 A0A182VIA4 A0A345AH67 A0A139WBY5 A0A182YGI2 A0A182L7I2 Q7PQN7 A0A182I7X3 A0A182RYI7 A0A182VSB6 A0A182P5Y9 A0A3B0JCV9 A0A182QAR7 A0A1I8NTF3 A0A182N5N3 A0A1A9W5L1 A0A182F6P1 B4MBR2 W5JC27 A0A1I8M4L5 A0A1I8M4K9 A0A1B0B4V9 A0A084VL74 A0A1A9YQ48 B4KB11 A0A1W4V450 A0A0R3NNU4 B4NHE1 E9GG50 A0A0M4F4U0 A0A0Q9VZP6 E9GBE9 A0A0P6CD76 A0A0Q9X5N6 A0A2P6LJY7 A0A0P6C9L9 A0A0P6HFJ0 T1IW42 A0A1D2N3W3 B3NYL4 E0VA32 A0A0P4Z0P0 W8C5G8 Q9VNM1-6 Q9VNM1 A0A0N7ZMC1 B4QXP8 Q9VNM1-4 A0A0B4K601 A0A0P5CAX0 A0A0P5YB18 A0A0P5WDL7 A0A0P5I5Y2 A0A0P5ZX39 A0A0P5MX59 A0A0N8DQ53 A0A164RXS6 A0A0P5MQL8 A0A0P6FUI5 A0A0P5WCS6 A0A0P5CZ61 A0A0R1E416 A0A0N8D8A5 B4PVM0 A0A0P4YK79 T1KR44
Pubmed
18725956
28756777
26354079
19121390
22118469
29403074
+ More
24845553 24130914 26483478 17510324 17994087 15632085 18362917 19820115 25244985 20966253 12364791 14747013 15626509 17210077 20920257 23761445 25315136 24438588 18057021 21292972 27289101 20566863 24495485 11897397 10731132 12537572 15677721 19837040 20164335 22422983 12537568 12537573 12537574 16110336 17569856 17569867 17550304
24845553 24130914 26483478 17510324 17994087 15632085 18362917 19820115 25244985 20966253 12364791 14747013 15626509 17210077 20920257 23761445 25315136 24438588 18057021 21292972 27289101 20566863 24495485 11897397 10731132 12537572 15677721 19837040 20164335 22422983 12537568 12537573 12537574 16110336 17569856 17569867 17550304
EMBL
AB330424
BAG68402.1
NWSH01002516
PCG68094.1
KT031001
ALM88299.1
+ More
PCG68093.1 KZ149905 PZC78337.1 KQ459144 KPJ03642.1 GDQN01008744 JAT82310.1 BABH01035606 AGBW02010970 OWR47452.1 GDQN01006862 JAT84192.1 KQ461188 KPJ07299.1 PYGN01000679 PSN42328.1 KK852623 KDR19960.1 NEVH01009379 PNF33311.1 DS231956 EDS28954.1 KC439539 AGX85007.1 JXUM01115799 JXUM01115800 JXUM01115801 KQ565916 KXJ70537.1 JXUM01054483 KQ561822 KXJ77427.1 CCAG010011761 CH477676 EAT37369.2 QCYY01002621 ROT68912.1 CH479179 EDW24275.1 CM000070 KRT00420.1 CH916377 EDV90724.1 EAL28070.3 KRT00421.1 KRT00423.1 MF175146 AXF35701.1 KQ971372 KYB25447.1 AY579078 AAAB01008880 AAT81602.1 EAA08566.3 APCN01002570 OUUW01000005 SPP80217.1 AXCN02001159 CH940656 EDW58533.2 ADMH02001637 ETN61661.1 JXJN01008559 ATLV01014424 KE524972 KFB38718.1 CH933806 EDW14689.1 KRG01147.1 KRT00422.1 CH964272 EDW84617.2 GL732543 EFX81337.1 CP012526 ALC46837.1 KRF78332.1 GL732538 EFX83169.1 GDIP01003024 JAN00692.1 KRG01146.1 MWRG01000011 PRD38886.1 GDIP01006624 LRGB01002860 JAM97091.1 KZS05899.1 GDIQ01019765 JAN74972.1 JH431607 LJIJ01000243 ODM99959.1 CH954181 EDV47995.1 DS235004 EEB10238.1 GDIP01219684 JAJ03718.1 GAMC01001762 JAC04794.1 AF364400 DQ666847 DQ666848 DQ666849 DQ666850 DQ666851 AE014297 AY071152 BT044552 AAF51909.3 AAK50050.1 AAL48774.1 ABH01175.1 GDIP01231369 JAI92032.1 CM000364 EDX11834.1 AFH06266.1 GDIP01202513 GDIP01201262 GDIP01173725 GDIP01123317 GDIP01095034 JAJ49677.1 GDIP01166693 GDIP01160573 GDIP01085406 GDIP01062604 GDIP01061044 JAM42671.1 GDIP01087610 JAM16105.1 GDIQ01217536 JAK34189.1 GDIP01235972 GDIP01208272 GDIP01200012 GDIP01155090 GDIP01121780 GDIP01038158 JAM65557.1 GDIQ01158311 JAK93414.1 GDIP01011823 JAM91892.1 LRGB01002121 KZS09045.1 GDIQ01160112 JAK91613.1 GDIQ01042849 JAN51888.1 GDIP01088852 JAM14863.1 GDIP01206670 GDIP01204670 GDIP01167620 GDIP01113120 GDIP01070803 JAJ55782.1 CM000160 KRK03990.1 KRK03991.1 KRK03992.1 GDIP01059007 JAM44708.1 EDW97829.1 GDIP01228480 JAI94921.1 CAEY01000383
PCG68093.1 KZ149905 PZC78337.1 KQ459144 KPJ03642.1 GDQN01008744 JAT82310.1 BABH01035606 AGBW02010970 OWR47452.1 GDQN01006862 JAT84192.1 KQ461188 KPJ07299.1 PYGN01000679 PSN42328.1 KK852623 KDR19960.1 NEVH01009379 PNF33311.1 DS231956 EDS28954.1 KC439539 AGX85007.1 JXUM01115799 JXUM01115800 JXUM01115801 KQ565916 KXJ70537.1 JXUM01054483 KQ561822 KXJ77427.1 CCAG010011761 CH477676 EAT37369.2 QCYY01002621 ROT68912.1 CH479179 EDW24275.1 CM000070 KRT00420.1 CH916377 EDV90724.1 EAL28070.3 KRT00421.1 KRT00423.1 MF175146 AXF35701.1 KQ971372 KYB25447.1 AY579078 AAAB01008880 AAT81602.1 EAA08566.3 APCN01002570 OUUW01000005 SPP80217.1 AXCN02001159 CH940656 EDW58533.2 ADMH02001637 ETN61661.1 JXJN01008559 ATLV01014424 KE524972 KFB38718.1 CH933806 EDW14689.1 KRG01147.1 KRT00422.1 CH964272 EDW84617.2 GL732543 EFX81337.1 CP012526 ALC46837.1 KRF78332.1 GL732538 EFX83169.1 GDIP01003024 JAN00692.1 KRG01146.1 MWRG01000011 PRD38886.1 GDIP01006624 LRGB01002860 JAM97091.1 KZS05899.1 GDIQ01019765 JAN74972.1 JH431607 LJIJ01000243 ODM99959.1 CH954181 EDV47995.1 DS235004 EEB10238.1 GDIP01219684 JAJ03718.1 GAMC01001762 JAC04794.1 AF364400 DQ666847 DQ666848 DQ666849 DQ666850 DQ666851 AE014297 AY071152 BT044552 AAF51909.3 AAK50050.1 AAL48774.1 ABH01175.1 GDIP01231369 JAI92032.1 CM000364 EDX11834.1 AFH06266.1 GDIP01202513 GDIP01201262 GDIP01173725 GDIP01123317 GDIP01095034 JAJ49677.1 GDIP01166693 GDIP01160573 GDIP01085406 GDIP01062604 GDIP01061044 JAM42671.1 GDIP01087610 JAM16105.1 GDIQ01217536 JAK34189.1 GDIP01235972 GDIP01208272 GDIP01200012 GDIP01155090 GDIP01121780 GDIP01038158 JAM65557.1 GDIQ01158311 JAK93414.1 GDIP01011823 JAM91892.1 LRGB01002121 KZS09045.1 GDIQ01160112 JAK91613.1 GDIQ01042849 JAN51888.1 GDIP01088852 JAM14863.1 GDIP01206670 GDIP01204670 GDIP01167620 GDIP01113120 GDIP01070803 JAJ55782.1 CM000160 KRK03990.1 KRK03991.1 KRK03992.1 GDIP01059007 JAM44708.1 EDW97829.1 GDIP01228480 JAI94921.1 CAEY01000383
Proteomes
UP000218220
UP000053268
UP000005204
UP000007151
UP000053240
UP000192223
+ More
UP000245037 UP000027135 UP000235965 UP000002320 UP000069940 UP000249989 UP000078200 UP000092444 UP000008820 UP000283509 UP000008744 UP000092445 UP000001819 UP000001070 UP000076407 UP000075903 UP000007266 UP000076408 UP000075882 UP000007062 UP000075840 UP000075900 UP000075920 UP000075885 UP000268350 UP000075886 UP000095300 UP000075884 UP000091820 UP000069272 UP000008792 UP000000673 UP000095301 UP000092460 UP000030765 UP000092443 UP000009192 UP000192221 UP000007798 UP000000305 UP000092553 UP000076858 UP000094527 UP000008711 UP000009046 UP000000803 UP000000304 UP000002282 UP000015104
UP000245037 UP000027135 UP000235965 UP000002320 UP000069940 UP000249989 UP000078200 UP000092444 UP000008820 UP000283509 UP000008744 UP000092445 UP000001819 UP000001070 UP000076407 UP000075903 UP000007266 UP000076408 UP000075882 UP000007062 UP000075840 UP000075900 UP000075920 UP000075885 UP000268350 UP000075886 UP000095300 UP000075884 UP000091820 UP000069272 UP000008792 UP000000673 UP000095301 UP000092460 UP000030765 UP000092443 UP000009192 UP000192221 UP000007798 UP000000305 UP000092553 UP000076858 UP000094527 UP000008711 UP000009046 UP000000803 UP000000304 UP000002282 UP000015104
Pfam
PF00001 7tm_1
ProteinModelPortal
B3XXL6
A0A2A4J983
A0A0S1YD64
A0A2A4J9L8
A0A2W1BYM3
A0A194QDU4
+ More
A0A1E1W5R9 H9IRZ6 A0A212F140 A0A1E1WB26 A0A0N1IES3 A0A1W4XJH0 A0A2P8YDJ1 A0A067RKH6 A0A2J7QXK0 A0A1S4FQV5 B0WJ64 U5N0V1 A0A182H7A0 A0A1A9VIV6 A0A182GCN7 A0A1B0ERV5 Q16SC4 A0A3R7PJV0 B4G380 A0A1A9ZT02 A0A0R3NHE1 B4JYG2 Q297Y5 A0A0R3NMC1 A0A0R3NH99 A0A182XIY4 A0A182VIA4 A0A345AH67 A0A139WBY5 A0A182YGI2 A0A182L7I2 Q7PQN7 A0A182I7X3 A0A182RYI7 A0A182VSB6 A0A182P5Y9 A0A3B0JCV9 A0A182QAR7 A0A1I8NTF3 A0A182N5N3 A0A1A9W5L1 A0A182F6P1 B4MBR2 W5JC27 A0A1I8M4L5 A0A1I8M4K9 A0A1B0B4V9 A0A084VL74 A0A1A9YQ48 B4KB11 A0A1W4V450 A0A0R3NNU4 B4NHE1 E9GG50 A0A0M4F4U0 A0A0Q9VZP6 E9GBE9 A0A0P6CD76 A0A0Q9X5N6 A0A2P6LJY7 A0A0P6C9L9 A0A0P6HFJ0 T1IW42 A0A1D2N3W3 B3NYL4 E0VA32 A0A0P4Z0P0 W8C5G8 Q9VNM1-6 Q9VNM1 A0A0N7ZMC1 B4QXP8 Q9VNM1-4 A0A0B4K601 A0A0P5CAX0 A0A0P5YB18 A0A0P5WDL7 A0A0P5I5Y2 A0A0P5ZX39 A0A0P5MX59 A0A0N8DQ53 A0A164RXS6 A0A0P5MQL8 A0A0P6FUI5 A0A0P5WCS6 A0A0P5CZ61 A0A0R1E416 A0A0N8D8A5 B4PVM0 A0A0P4YK79 T1KR44
A0A1E1W5R9 H9IRZ6 A0A212F140 A0A1E1WB26 A0A0N1IES3 A0A1W4XJH0 A0A2P8YDJ1 A0A067RKH6 A0A2J7QXK0 A0A1S4FQV5 B0WJ64 U5N0V1 A0A182H7A0 A0A1A9VIV6 A0A182GCN7 A0A1B0ERV5 Q16SC4 A0A3R7PJV0 B4G380 A0A1A9ZT02 A0A0R3NHE1 B4JYG2 Q297Y5 A0A0R3NMC1 A0A0R3NH99 A0A182XIY4 A0A182VIA4 A0A345AH67 A0A139WBY5 A0A182YGI2 A0A182L7I2 Q7PQN7 A0A182I7X3 A0A182RYI7 A0A182VSB6 A0A182P5Y9 A0A3B0JCV9 A0A182QAR7 A0A1I8NTF3 A0A182N5N3 A0A1A9W5L1 A0A182F6P1 B4MBR2 W5JC27 A0A1I8M4L5 A0A1I8M4K9 A0A1B0B4V9 A0A084VL74 A0A1A9YQ48 B4KB11 A0A1W4V450 A0A0R3NNU4 B4NHE1 E9GG50 A0A0M4F4U0 A0A0Q9VZP6 E9GBE9 A0A0P6CD76 A0A0Q9X5N6 A0A2P6LJY7 A0A0P6C9L9 A0A0P6HFJ0 T1IW42 A0A1D2N3W3 B3NYL4 E0VA32 A0A0P4Z0P0 W8C5G8 Q9VNM1-6 Q9VNM1 A0A0N7ZMC1 B4QXP8 Q9VNM1-4 A0A0B4K601 A0A0P5CAX0 A0A0P5YB18 A0A0P5WDL7 A0A0P5I5Y2 A0A0P5ZX39 A0A0P5MX59 A0A0N8DQ53 A0A164RXS6 A0A0P5MQL8 A0A0P6FUI5 A0A0P5WCS6 A0A0P5CZ61 A0A0R1E416 A0A0N8D8A5 B4PVM0 A0A0P4YK79 T1KR44
PDB
5ZBQ
E-value=4.68799e-29,
Score=318
Ontologies
GO
Topology
Subcellular location
Membrane
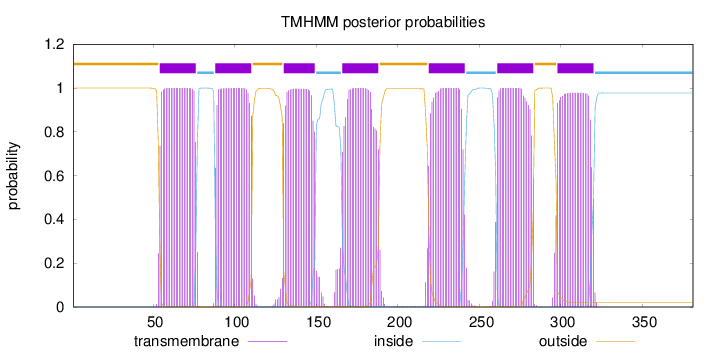
Length:
381
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
155.24053
Exp number, first 60 AAs:
6.96999
Total prob of N-in:
0.00011
outside
1 - 53
TMhelix
54 - 76
inside
77 - 87
TMhelix
88 - 110
outside
111 - 129
TMhelix
130 - 149
inside
150 - 165
TMhelix
166 - 188
outside
189 - 218
TMhelix
219 - 241
inside
242 - 260
TMhelix
261 - 283
outside
284 - 297
TMhelix
298 - 320
inside
321 - 381
Population Genetic Test Statistics
Pi
180.47584
Theta
174.573911
Tajima's D
0.302787
CLR
18.80785
CSRT
0.454427278636068
Interpretation
Uncertain
