Gene
KWMTBOMO15802
Annotation
uncharacterized_LOC106116239_precursor_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.729
Sequence
CDS
ATGTATCAAGCGAGATGGCTGATGCTGGCGGTGGCAGCGACGTTAGCGCTGCAGTCGCGAGGGGATGGCGGCATCAGACTACCAGACCAGCCTTCCAAAGAAAACACCGAAGGAAGATCACTCAATTTCGGAGGAGATAATGGAAGATCCCCTCAGGCTCAGGGTCGTGGTTTACTGGACTGGATTGGCTTGGGCGAGGATCAAGACCCCTACATCCAGCAATCAACACAGCAGTGCATCAATGGAGACCTAGCAGACTGTTTCAAAGCACAAGCTTTGAGGTCGTTTGACGATTTCTTCGACAAGCAGGCTTACCAACTTTCAGATAACGCACTCTTAGTCAAAGCAGAGAACCAAGCAAGGGCACTAGCCAGAGAGCCACCTCAACTGGATTCGCAGCCTCGTAGCGAAGATTCCGACTGGGAAGCCCTTGTCAAATACGGAATGAGGAAAATTGAAAGGTTTTTAAGGTCAACCGCCTTGGAGCTGCAGCTTGACAATGAAGTGACTTCCGAGGGTGTTATCGCTCCGAGATTCATCGACGAGATTAATGATGAAGTAGACGTAATCGAGGATAAAACGGCGCCGTTCCGTCGCCACAAGCTGAAGAAACTTATCATTCCTATGCTACTCATCCTCAAGTTGTTCAAGCTGAAACTGCTTCTCTTCCTGCCTCTCATTCTTGGTTTGGCCAGCTTCAAGAAATTCTTAGGATTCATGGCTCTTATCATTCCAGGTGTAATCGGATATTTCAAATTCTGCAAGCCAAATTCATCTCCTTTCTCCACAAACCACTATACGGGGCCTCAGTATTCACCCGCCGGTATTGGTTTAGGACCTTACAGAGAAAGTCCTCCCGAAAACTATGGCCATTACAATAATTATCCCGGCGGCCACAAGTACAGCCAGGGGGGCTTTTCGTTCAGAGACCACGACGCCAGTCCCCAAGACCTGGCCTATCAAGGCTGGGAGTACAGGAACAAGAAAGGAGCTGAAGACATCAAGGCTGAGAGCCCATAA
Protein
MYQARWLMLAVAATLALQSRGDGGIRLPDQPSKENTEGRSLNFGGDNGRSPQAQGRGLLDWIGLGEDQDPYIQQSTQQCINGDLADCFKAQALRSFDDFFDKQAYQLSDNALLVKAENQARALAREPPQLDSQPRSEDSDWEALVKYGMRKIERFLRSTALELQLDNEVTSEGVIAPRFIDEINDEVDVIEDKTAPFRRHKLKKLIIPMLLILKLFKLKLLLFLPLILGLASFKKFLGFMALIIPGVIGYFKFCKPNSSPFSTNHYTGPQYSPAGIGLGPYRESPPENYGHYNNYPGGHKYSQGGFSFRDHDASPQDLAYQGWEYRNKKGAEDIKAESP
Summary
Uniprot
I4DJ13
A0A0N1IBE3
A0A2W1BD43
A0A2A4JZB7
A0A194QDZ9
I4DMQ8
+ More
A0A2H1VLY7 A0A212F3S2 A0A3S2TQZ1 A0A0L7KW43 N6UC33 D6X0U7 B0WLW8 A0A087Z2Q4 W5JK00 A0A182F6P3 A0A0T6BAE8 A0A182K0G1 A0A182P5Y7 A0A182YGI4 A0A1W4XCN4 A0A182RYI9 A0A182I7X1 A0A182VSB8 Q7QB96 A0A182VMR1 A0A182XIY6 A0A182QMA0 A0A182T899 A0A182U9X3 A0A182MA36 A0A084VL79 A0A182N5N5 A0A182GKT6 A0A182JIU9 A0A232EU02 K7IU75 A0A0C9QUH2 A0A067R8W6 A0A2J7PI35 A0A154PH10 A0A088AGF2 E0VA34 A0A0L7RJD9 A0A0M9A9V5 A0A310SN67 Q16SC8 A0A087ZXZ9 A0A1S4FRB5 A0A2A3EPH8 A0A1J1HVQ7 A0A034WR25 W8C419 A0A1A9VIV7 A0A1A9YQ44 A0A034WTJ0 A0A0C9RYD7 A0A1B0B4V6 A0A1B6EFN1 B4MBR0 A0A336MM41 A0A336MHL8 A0A1A9ZSY5 B4NHD9 B4KB14 B3LVV5 B4G382 A0A0A1WY92 Q297Y7 A0A1B0G0J7 A0A0M4EQZ0 A0A0L0BMQ7 A0A195FI59 W8BPB5 B3NYL6 A0A3B0JDS1 B4I4C2 B4QY80 Q9VNM3 A0A026WHV4 B4PVM2 A0A182L7H8 E2APJ3 A0A2S2PLG6 A0A0J7NUG7 A0A1I8MVA1 J9K4V3 A0A1W4URW8 B4JYG0 A0A195B157 F4X746 A0A195DIZ7 A0A158NDA1 E2B2W2 A0A336MK83
A0A2H1VLY7 A0A212F3S2 A0A3S2TQZ1 A0A0L7KW43 N6UC33 D6X0U7 B0WLW8 A0A087Z2Q4 W5JK00 A0A182F6P3 A0A0T6BAE8 A0A182K0G1 A0A182P5Y7 A0A182YGI4 A0A1W4XCN4 A0A182RYI9 A0A182I7X1 A0A182VSB8 Q7QB96 A0A182VMR1 A0A182XIY6 A0A182QMA0 A0A182T899 A0A182U9X3 A0A182MA36 A0A084VL79 A0A182N5N5 A0A182GKT6 A0A182JIU9 A0A232EU02 K7IU75 A0A0C9QUH2 A0A067R8W6 A0A2J7PI35 A0A154PH10 A0A088AGF2 E0VA34 A0A0L7RJD9 A0A0M9A9V5 A0A310SN67 Q16SC8 A0A087ZXZ9 A0A1S4FRB5 A0A2A3EPH8 A0A1J1HVQ7 A0A034WR25 W8C419 A0A1A9VIV7 A0A1A9YQ44 A0A034WTJ0 A0A0C9RYD7 A0A1B0B4V6 A0A1B6EFN1 B4MBR0 A0A336MM41 A0A336MHL8 A0A1A9ZSY5 B4NHD9 B4KB14 B3LVV5 B4G382 A0A0A1WY92 Q297Y7 A0A1B0G0J7 A0A0M4EQZ0 A0A0L0BMQ7 A0A195FI59 W8BPB5 B3NYL6 A0A3B0JDS1 B4I4C2 B4QY80 Q9VNM3 A0A026WHV4 B4PVM2 A0A182L7H8 E2APJ3 A0A2S2PLG6 A0A0J7NUG7 A0A1I8MVA1 J9K4V3 A0A1W4URW8 B4JYG0 A0A195B157 F4X746 A0A195DIZ7 A0A158NDA1 E2B2W2 A0A336MK83
Pubmed
22651552
26354079
28756777
22118469
26227816
23537049
+ More
18362917 19820115 20920257 25244985 12364791 14747013 17210077 24438588 26483478 28648823 20075255 24845553 20566863 17510324 25348373 24495485 17994087 25830018 15632085 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 30249741 17550304 20966253 20798317 25315136 21719571 21347285
18362917 19820115 20920257 25244985 12364791 14747013 17210077 24438588 26483478 28648823 20075255 24845553 20566863 17510324 25348373 24495485 17994087 25830018 15632085 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 30249741 17550304 20966253 20798317 25315136 21719571 21347285
EMBL
AK401281
BAM17903.1
KQ461188
KPJ07306.1
KZ150351
PZC71257.1
+ More
NWSH01000392 PCG76732.1 KQ459144 KPJ03649.1 AK402576 BAM19198.1 ODYU01003110 SOQ41452.1 AGBW02010496 OWR48387.1 RSAL01000019 RVE52737.1 JTDY01005182 KOB67286.1 APGK01029581 APGK01029582 KB740735 KB631809 ENN79215.1 ERL86306.1 KQ971372 EFA09547.1 DS231991 EDS30703.1 ADMH02000905 ETN64707.1 LJIG01002643 KRT84297.1 APCN01002567 AAAB01008880 EAA08637.4 AXCN02001159 AXCM01003891 ATLV01014426 KE524972 KFB38723.1 JXUM01070687 KQ562622 KXJ75475.1 NNAY01002222 OXU21776.1 AAZX01005587 GBYB01004282 GBYB01004283 JAG74049.1 JAG74050.1 KK852623 KDR19943.1 NEVH01025129 PNF15984.1 KQ434902 KZC11145.1 DS235004 EEB10240.1 KQ414581 KOC70949.1 KQ435706 KOX80170.1 KQ759821 OAD62705.1 CH477676 EAT37365.1 KZ288200 PBC33600.1 CVRI01000023 CRK92101.1 GAKP01001858 JAC57094.1 GAMC01005794 JAC00762.1 GAKP01001859 JAC57093.1 GBYB01014265 JAG84032.1 JXJN01008558 GEDC01000560 JAS36738.1 CH940656 EDW58531.1 UFQT01001300 SSX29903.1 SSX29902.1 CH964272 EDW84615.1 CH933806 EDW14692.1 CH902617 EDV43729.1 CH479179 EDW24277.1 GBXI01010248 JAD04044.1 CM000070 EAL28068.2 CCAG010011761 CP012526 ALC46835.1 JRES01001623 KNC21370.1 KQ981560 KYN39942.1 GAMC01005793 JAC00763.1 CH954181 EDV47997.1 OUUW01000005 SPP80215.1 CH480821 EDW55065.1 CM000364 EDX11837.1 AE014297 BT015251 AAF51907.2 AAN13234.1 AAT94480.1 AGB95686.1 KK107199 QOIP01000009 EZA55583.1 RLU18963.1 CM000160 EDW97831.1 KRK03994.1 GL441542 EFN64649.1 GGMR01017671 MBY30290.1 LBMM01001591 KMQ96050.1 ABLF02016062 CH916377 EDV90722.1 KQ976690 KYM78027.1 GL888828 EGI57658.1 KQ980800 KYN12863.1 ADTU01012534 GL445250 EFN89931.1 SSX29901.1
NWSH01000392 PCG76732.1 KQ459144 KPJ03649.1 AK402576 BAM19198.1 ODYU01003110 SOQ41452.1 AGBW02010496 OWR48387.1 RSAL01000019 RVE52737.1 JTDY01005182 KOB67286.1 APGK01029581 APGK01029582 KB740735 KB631809 ENN79215.1 ERL86306.1 KQ971372 EFA09547.1 DS231991 EDS30703.1 ADMH02000905 ETN64707.1 LJIG01002643 KRT84297.1 APCN01002567 AAAB01008880 EAA08637.4 AXCN02001159 AXCM01003891 ATLV01014426 KE524972 KFB38723.1 JXUM01070687 KQ562622 KXJ75475.1 NNAY01002222 OXU21776.1 AAZX01005587 GBYB01004282 GBYB01004283 JAG74049.1 JAG74050.1 KK852623 KDR19943.1 NEVH01025129 PNF15984.1 KQ434902 KZC11145.1 DS235004 EEB10240.1 KQ414581 KOC70949.1 KQ435706 KOX80170.1 KQ759821 OAD62705.1 CH477676 EAT37365.1 KZ288200 PBC33600.1 CVRI01000023 CRK92101.1 GAKP01001858 JAC57094.1 GAMC01005794 JAC00762.1 GAKP01001859 JAC57093.1 GBYB01014265 JAG84032.1 JXJN01008558 GEDC01000560 JAS36738.1 CH940656 EDW58531.1 UFQT01001300 SSX29903.1 SSX29902.1 CH964272 EDW84615.1 CH933806 EDW14692.1 CH902617 EDV43729.1 CH479179 EDW24277.1 GBXI01010248 JAD04044.1 CM000070 EAL28068.2 CCAG010011761 CP012526 ALC46835.1 JRES01001623 KNC21370.1 KQ981560 KYN39942.1 GAMC01005793 JAC00763.1 CH954181 EDV47997.1 OUUW01000005 SPP80215.1 CH480821 EDW55065.1 CM000364 EDX11837.1 AE014297 BT015251 AAF51907.2 AAN13234.1 AAT94480.1 AGB95686.1 KK107199 QOIP01000009 EZA55583.1 RLU18963.1 CM000160 EDW97831.1 KRK03994.1 GL441542 EFN64649.1 GGMR01017671 MBY30290.1 LBMM01001591 KMQ96050.1 ABLF02016062 CH916377 EDV90722.1 KQ976690 KYM78027.1 GL888828 EGI57658.1 KQ980800 KYN12863.1 ADTU01012534 GL445250 EFN89931.1 SSX29901.1
Proteomes
UP000053240
UP000218220
UP000053268
UP000007151
UP000283053
UP000037510
+ More
UP000019118 UP000030742 UP000007266 UP000002320 UP000000673 UP000069272 UP000075881 UP000075885 UP000076408 UP000192223 UP000075900 UP000075840 UP000075920 UP000007062 UP000075903 UP000076407 UP000075886 UP000075901 UP000075902 UP000075883 UP000030765 UP000075884 UP000069940 UP000249989 UP000075880 UP000215335 UP000002358 UP000027135 UP000235965 UP000076502 UP000005203 UP000009046 UP000053825 UP000053105 UP000008820 UP000242457 UP000183832 UP000078200 UP000092443 UP000092460 UP000008792 UP000092445 UP000007798 UP000009192 UP000007801 UP000008744 UP000001819 UP000092444 UP000092553 UP000037069 UP000078541 UP000008711 UP000268350 UP000001292 UP000000304 UP000000803 UP000053097 UP000279307 UP000002282 UP000075882 UP000000311 UP000036403 UP000095301 UP000007819 UP000192221 UP000001070 UP000078540 UP000007755 UP000078492 UP000005205 UP000008237
UP000019118 UP000030742 UP000007266 UP000002320 UP000000673 UP000069272 UP000075881 UP000075885 UP000076408 UP000192223 UP000075900 UP000075840 UP000075920 UP000007062 UP000075903 UP000076407 UP000075886 UP000075901 UP000075902 UP000075883 UP000030765 UP000075884 UP000069940 UP000249989 UP000075880 UP000215335 UP000002358 UP000027135 UP000235965 UP000076502 UP000005203 UP000009046 UP000053825 UP000053105 UP000008820 UP000242457 UP000183832 UP000078200 UP000092443 UP000092460 UP000008792 UP000092445 UP000007798 UP000009192 UP000007801 UP000008744 UP000001819 UP000092444 UP000092553 UP000037069 UP000078541 UP000008711 UP000268350 UP000001292 UP000000304 UP000000803 UP000053097 UP000279307 UP000002282 UP000075882 UP000000311 UP000036403 UP000095301 UP000007819 UP000192221 UP000001070 UP000078540 UP000007755 UP000078492 UP000005205 UP000008237
PRIDE
Pfam
PF07898 DUF1676
Interpro
IPR012464
DUF1676
ProteinModelPortal
I4DJ13
A0A0N1IBE3
A0A2W1BD43
A0A2A4JZB7
A0A194QDZ9
I4DMQ8
+ More
A0A2H1VLY7 A0A212F3S2 A0A3S2TQZ1 A0A0L7KW43 N6UC33 D6X0U7 B0WLW8 A0A087Z2Q4 W5JK00 A0A182F6P3 A0A0T6BAE8 A0A182K0G1 A0A182P5Y7 A0A182YGI4 A0A1W4XCN4 A0A182RYI9 A0A182I7X1 A0A182VSB8 Q7QB96 A0A182VMR1 A0A182XIY6 A0A182QMA0 A0A182T899 A0A182U9X3 A0A182MA36 A0A084VL79 A0A182N5N5 A0A182GKT6 A0A182JIU9 A0A232EU02 K7IU75 A0A0C9QUH2 A0A067R8W6 A0A2J7PI35 A0A154PH10 A0A088AGF2 E0VA34 A0A0L7RJD9 A0A0M9A9V5 A0A310SN67 Q16SC8 A0A087ZXZ9 A0A1S4FRB5 A0A2A3EPH8 A0A1J1HVQ7 A0A034WR25 W8C419 A0A1A9VIV7 A0A1A9YQ44 A0A034WTJ0 A0A0C9RYD7 A0A1B0B4V6 A0A1B6EFN1 B4MBR0 A0A336MM41 A0A336MHL8 A0A1A9ZSY5 B4NHD9 B4KB14 B3LVV5 B4G382 A0A0A1WY92 Q297Y7 A0A1B0G0J7 A0A0M4EQZ0 A0A0L0BMQ7 A0A195FI59 W8BPB5 B3NYL6 A0A3B0JDS1 B4I4C2 B4QY80 Q9VNM3 A0A026WHV4 B4PVM2 A0A182L7H8 E2APJ3 A0A2S2PLG6 A0A0J7NUG7 A0A1I8MVA1 J9K4V3 A0A1W4URW8 B4JYG0 A0A195B157 F4X746 A0A195DIZ7 A0A158NDA1 E2B2W2 A0A336MK83
A0A2H1VLY7 A0A212F3S2 A0A3S2TQZ1 A0A0L7KW43 N6UC33 D6X0U7 B0WLW8 A0A087Z2Q4 W5JK00 A0A182F6P3 A0A0T6BAE8 A0A182K0G1 A0A182P5Y7 A0A182YGI4 A0A1W4XCN4 A0A182RYI9 A0A182I7X1 A0A182VSB8 Q7QB96 A0A182VMR1 A0A182XIY6 A0A182QMA0 A0A182T899 A0A182U9X3 A0A182MA36 A0A084VL79 A0A182N5N5 A0A182GKT6 A0A182JIU9 A0A232EU02 K7IU75 A0A0C9QUH2 A0A067R8W6 A0A2J7PI35 A0A154PH10 A0A088AGF2 E0VA34 A0A0L7RJD9 A0A0M9A9V5 A0A310SN67 Q16SC8 A0A087ZXZ9 A0A1S4FRB5 A0A2A3EPH8 A0A1J1HVQ7 A0A034WR25 W8C419 A0A1A9VIV7 A0A1A9YQ44 A0A034WTJ0 A0A0C9RYD7 A0A1B0B4V6 A0A1B6EFN1 B4MBR0 A0A336MM41 A0A336MHL8 A0A1A9ZSY5 B4NHD9 B4KB14 B3LVV5 B4G382 A0A0A1WY92 Q297Y7 A0A1B0G0J7 A0A0M4EQZ0 A0A0L0BMQ7 A0A195FI59 W8BPB5 B3NYL6 A0A3B0JDS1 B4I4C2 B4QY80 Q9VNM3 A0A026WHV4 B4PVM2 A0A182L7H8 E2APJ3 A0A2S2PLG6 A0A0J7NUG7 A0A1I8MVA1 J9K4V3 A0A1W4URW8 B4JYG0 A0A195B157 F4X746 A0A195DIZ7 A0A158NDA1 E2B2W2 A0A336MK83
Ontologies
KEGG
PANTHER
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.991070
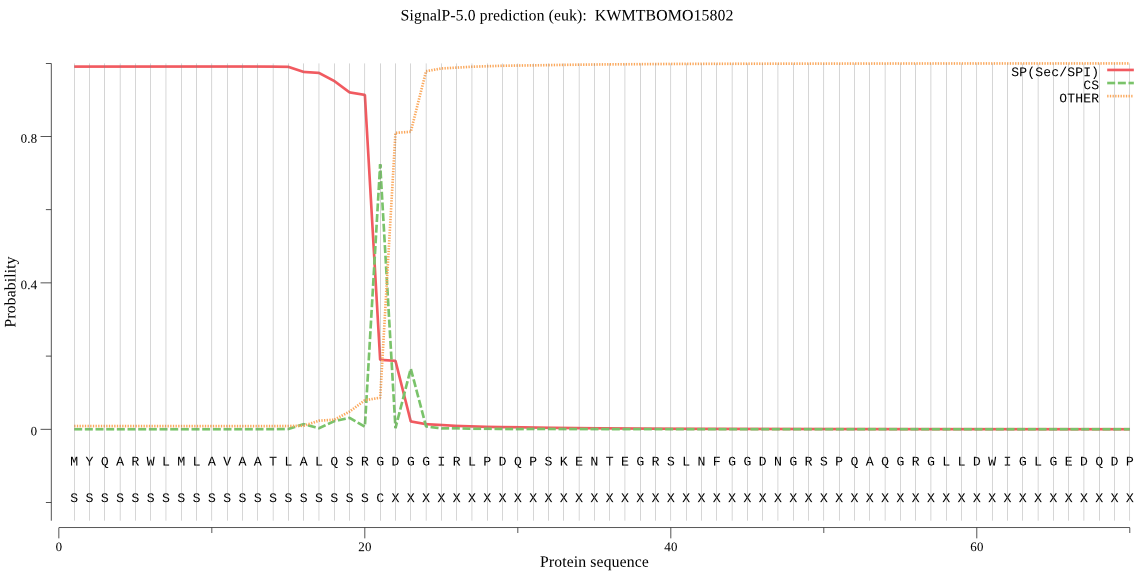
Length:
339
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
36.80914
Exp number, first 60 AAs:
0.01302
Total prob of N-in:
0.78350
inside
1 - 204
TMhelix
205 - 227
outside
228 - 236
TMhelix
237 - 254
inside
255 - 339

Population Genetic Test Statistics
Pi
204.420354
Theta
175.530649
Tajima's D
0.353186
CLR
0.836485
CSRT
0.469626518674066
Interpretation
Uncertain
