Gene
KWMTBOMO15797 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000028
Annotation
PREDICTED:_uncharacterized_protein_LOC105390186_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 1.741
Sequence
CDS
ATGACGAGACTCAGCATAATAGCAATAGTGCTATGCCAGATATTGTGCTTGCAAATCCAGGGCGGGCACACCGAAGATGATAACCCTCTGCTGGATATCGCGTCGTCATTGCTCCAAAACATGGGCAATGGCGACAACAATGGCAACGGAATGGCGGCCATTGGAAGTATTATCGGAAACCTCATGCAGGGTGACAACGTTAAGAACTTGGCATCTTTATTTAACAATGAAAAAGAGGAAAAAGAAAAAGATAGCGGCAACATCGGAGATATTCTTTCAGGCTTAGGCAGTTTGATGGGCGGACAAGACGGAAAAATCGATCCAGCCATGATCGGAGGAATGATCTCGATGTTCGCCTCCATGGGTAGCACGCCTAAACGCGAGAAAAGGGAACAGAAAAAGGAAATAACCTTCGACAATTTAATGAACTTAGCATCGAGCTTCACGCAAAACAAAGAAGGCGGTAGCTACATGCCGCTAATCATGAGTGCTTTGAAAGGGTTCACGAAAATGGAAGCCGATAAGAAAGCCGACGAACACAAGGACCACGCATCCTTCCTGCCGCCGTACCTAGAAAAAGCGCATCTATACTGGGACATATTCATAAACTCCGAAGTCGGCAAACTGGTTTGGGAGAAAACCGGCATGAAGAAAATGTTCAAAGCCTTCACCGGCCCCGACGGGAAGATCAGCTTCGAAACTATGTTCAAGAACTTCGAGAATACGTCGTTCAGAAGACATTGGATTAAGGCCTCCGCCAAATATTTGACCGACATGGCTGTTCATGTGACGAAGCCTGAAGTATATAGGAGGTACATATCGATGGCGGAATACGTTATCAACTCGTTCATGGAGTCCCAAGGATTCCCTAACAGCATACAATTCAACGCCAAAGCTCCGGCTAAATCGCTAACGGCCATCATCAATTACCTTCTGAAGACTTACATGGACTTCGATGCTGACGTTACCGAGTACGTCGTCCCTGCGGTCGAATACGCCAAGCAAACGTTGAAGCTAGCGGAGAAGGCCGCCCAGTCTGTGGCCACCCGTGAAGACTACGCCGCCGTCAGCGACCGCCTCACGGACGCGCTCAACCTCGAGGTCATCGAGCCGGTGCTGCGGGTGTACCGCGCCTACCGGCACTCCGCCGCCGCGCCGCACTGCCAGGAGCACCTCATGTGTCTCGTCAACCGGCCGGAGGGCGACAGGAAAGGAGCCCCGGGTCTCAAAGCCGGACTGACGAAGCTCAGCAGCTTAGTGGCCTCCGCGGCCCTCAGCTTCCACGACGGAAAAGGATTCTGGGACCTCTACAACGCGATCCAGAGCGACGTCAATTGCGAGGCCGCCTACCCCGCCGACTGCTCCGCCTTCCACGAGCACGAACTCAGAGTCACCACGGAGCCCTACCACACTGAACTATAA
Protein
MTRLSIIAIVLCQILCLQIQGGHTEDDNPLLDIASSLLQNMGNGDNNGNGMAAIGSIIGNLMQGDNVKNLASLFNNEKEEKEKDSGNIGDILSGLGSLMGGQDGKIDPAMIGGMISMFASMGSTPKREKREQKKEITFDNLMNLASSFTQNKEGGSYMPLIMSALKGFTKMEADKKADEHKDHASFLPPYLEKAHLYWDIFINSEVGKLVWEKTGMKKMFKAFTGPDGKISFETMFKNFENTSFRRHWIKASAKYLTDMAVHVTKPEVYRRYISMAEYVINSFMESQGFPNSIQFNAKAPAKSLTAIINYLLKTYMDFDADVTEYVVPAVEYAKQTLKLAEKAAQSVATREDYAAVSDRLTDALNLEVIEPVLRVYRAYRHSAAAPHCQEHLMCLVNRPEGDRKGAPGLKAGLTKLSSLVASAALSFHDGKGFWDLYNAIQSDVNCEAAYPADCSAFHEHELRVTTEPYHTEL
Summary
Uniprot
H9IS05
A0A2H1VAF6
A0A2A4JEN5
A0A2W1BC70
I4DMH0
A0A0N1INB0
+ More
A0A212FBG2 A0A194QDV3 A0A0L7KVA7 A0A1J1J651 A0A1S4F843 Q17CK1 A0A1B0G226 A0A182GGR3 A0A0L0BRC1 A0A336KMX4 A0A1I8MVK0 V5GIN9 A0A1A9VDQ1 A0A1I8QCX6 A0A1B0B2Y7 Q9VHH1 A0A182KB23 A0A182HYP1 A0A182KZI9 Q7QA05 A0A182XII8 A0A182TWG5 A0A182VA19 A0A0K8WC62 A0A1W4VQR3 A0A182PP77 A0A182RE42 A0A3B0JDK2 Q29CF9 A0A1A9W5N9 K7IPI4 A0A034VUM8 A0A182SQ60 A0A182MMQ6 A0A182Y8S4 A0A0K8UB59 B3M1L9 A0A182NGF6 B3P1T0 A0A182Q8G0 A0A182IP29 A0A1A9Y0J1 A0A0M4EQR6 B4PU09 A0A0J7NXA9 B4QX91 B4GN40 W5J9J2 B4NG19 A0A084WP51 B4JRK2 A0A2J7PYT1 A0A182F6M5 B4M0P5 B4KAN8 A0A026WBX3 A0A0A1WPG8 A0A182W3F9 A0A0L7QKX4 A0A088AFD1 D6X2F8 J9K5M4 E2AS40 A0A2A3EUV6 A0A2S2NTR6 A0A151WQS4 A0A154PST5 A0A195E4I8 A0A195FGG5 A0A158NXZ2 A0A195BS98 E9ICC8 F4X4D2 E2B432 A0A1B6JSR3 N6UF93 A0A2S2Q674 A0A151IGQ5 V9PB56 A0A1A9ZSW4 A0A067R2A3 A0A1W4X0J0 B0X1K0 A0A2P8ZP51 R4WP12 A0A1Y1LD01 A0A0K8UV50
A0A212FBG2 A0A194QDV3 A0A0L7KVA7 A0A1J1J651 A0A1S4F843 Q17CK1 A0A1B0G226 A0A182GGR3 A0A0L0BRC1 A0A336KMX4 A0A1I8MVK0 V5GIN9 A0A1A9VDQ1 A0A1I8QCX6 A0A1B0B2Y7 Q9VHH1 A0A182KB23 A0A182HYP1 A0A182KZI9 Q7QA05 A0A182XII8 A0A182TWG5 A0A182VA19 A0A0K8WC62 A0A1W4VQR3 A0A182PP77 A0A182RE42 A0A3B0JDK2 Q29CF9 A0A1A9W5N9 K7IPI4 A0A034VUM8 A0A182SQ60 A0A182MMQ6 A0A182Y8S4 A0A0K8UB59 B3M1L9 A0A182NGF6 B3P1T0 A0A182Q8G0 A0A182IP29 A0A1A9Y0J1 A0A0M4EQR6 B4PU09 A0A0J7NXA9 B4QX91 B4GN40 W5J9J2 B4NG19 A0A084WP51 B4JRK2 A0A2J7PYT1 A0A182F6M5 B4M0P5 B4KAN8 A0A026WBX3 A0A0A1WPG8 A0A182W3F9 A0A0L7QKX4 A0A088AFD1 D6X2F8 J9K5M4 E2AS40 A0A2A3EUV6 A0A2S2NTR6 A0A151WQS4 A0A154PST5 A0A195E4I8 A0A195FGG5 A0A158NXZ2 A0A195BS98 E9ICC8 F4X4D2 E2B432 A0A1B6JSR3 N6UF93 A0A2S2Q674 A0A151IGQ5 V9PB56 A0A1A9ZSW4 A0A067R2A3 A0A1W4X0J0 B0X1K0 A0A2P8ZP51 R4WP12 A0A1Y1LD01 A0A0K8UV50
Pubmed
19121390
28756777
22651552
26354079
22118469
26227816
+ More
17510324 26483478 26108605 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 12364791 14747013 17210077 15632085 17994087 20075255 25348373 25244985 17550304 20920257 23761445 24438588 24508170 30249741 25830018 18362917 19820115 20798317 21347285 21282665 21719571 23537049 24845553 29403074 23691247 28004739
17510324 26483478 26108605 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 12364791 14747013 17210077 15632085 17994087 20075255 25348373 25244985 17550304 20920257 23761445 24438588 24508170 30249741 25830018 18362917 19820115 20798317 21347285 21282665 21719571 23537049 24845553 29403074 23691247 28004739
EMBL
BABH01035561
ODYU01001334
SOQ37372.1
NWSH01001860
PCG69883.1
KZ150146
+ More
PZC72889.1 AK402488 BAM19110.1 KQ461188 KPJ07309.1 AGBW02009344 OWR51058.1 KQ459144 KPJ03652.1 JTDY01005255 KOB67192.1 CVRI01000072 CRL07474.1 CH477308 EAT44020.1 CCAG010010758 JXUM01062409 JXUM01062410 JXUM01062411 JXUM01062412 KQ562198 KXJ76431.1 JRES01001482 KNC22533.1 UFQS01000283 UFQT01000283 SSX02400.1 SSX22775.1 GALX01004552 JAB63914.1 JXJN01007739 AE014297 AY051945 AAF54343.1 AAK93369.1 APCN01005395 AAAB01008898 EAA09114.4 GDHF01003874 JAI48440.1 OUUW01000005 SPP80165.1 CM000070 EAL26687.2 GAKP01013744 JAC45208.1 AXCM01001003 GDHF01028407 JAI23907.1 CH902617 EDV43310.1 CH954181 EDV49818.1 AXCN02001608 CP012526 ALC47388.1 CM000160 EDW96620.1 KRK03243.1 KRK03244.1 KRK03245.1 KRK03246.1 LBMM01001020 KMQ97055.1 CM000364 EDX13662.1 CH479186 EDW39166.1 ADMH02001987 ETN60078.1 CH964251 EDW83236.1 ATLV01024906 KE525364 KFB51995.1 CH916373 EDV94392.1 NEVH01020342 PNF21460.1 CH940650 EDW67337.1 CH933806 EDW16775.1 KK107323 QOIP01000002 EZA52559.1 RLU26175.1 GBXI01013550 JAD00742.1 KQ414940 KOC59235.1 KQ971372 EFA10266.1 ABLF02036082 GL442221 EFN63740.1 KZ288185 PBC34929.1 GGMR01007898 MBY20517.1 KQ982815 KYQ50216.1 KQ435180 KZC14971.1 KQ979685 KYN19774.1 KQ981606 KYN39488.1 ADTU01000542 KQ976423 KYM88890.1 GL762231 EFZ21778.1 GL888633 EGI58728.1 GL445463 EFN89546.1 GECU01005462 JAT02245.1 APGK01024354 APGK01024355 KB740544 KB632130 ENN80410.1 ERL88961.1 GGMS01004011 MBY73214.1 KQ977671 KYN00619.1 KC282411 AGX25168.1 KK852806 KDR16096.1 DS232261 EDS38649.1 PYGN01000005 PSN58281.1 AK417411 BAN20626.1 GEZM01062872 JAV69446.1 GDHF01021740 JAI30574.1
PZC72889.1 AK402488 BAM19110.1 KQ461188 KPJ07309.1 AGBW02009344 OWR51058.1 KQ459144 KPJ03652.1 JTDY01005255 KOB67192.1 CVRI01000072 CRL07474.1 CH477308 EAT44020.1 CCAG010010758 JXUM01062409 JXUM01062410 JXUM01062411 JXUM01062412 KQ562198 KXJ76431.1 JRES01001482 KNC22533.1 UFQS01000283 UFQT01000283 SSX02400.1 SSX22775.1 GALX01004552 JAB63914.1 JXJN01007739 AE014297 AY051945 AAF54343.1 AAK93369.1 APCN01005395 AAAB01008898 EAA09114.4 GDHF01003874 JAI48440.1 OUUW01000005 SPP80165.1 CM000070 EAL26687.2 GAKP01013744 JAC45208.1 AXCM01001003 GDHF01028407 JAI23907.1 CH902617 EDV43310.1 CH954181 EDV49818.1 AXCN02001608 CP012526 ALC47388.1 CM000160 EDW96620.1 KRK03243.1 KRK03244.1 KRK03245.1 KRK03246.1 LBMM01001020 KMQ97055.1 CM000364 EDX13662.1 CH479186 EDW39166.1 ADMH02001987 ETN60078.1 CH964251 EDW83236.1 ATLV01024906 KE525364 KFB51995.1 CH916373 EDV94392.1 NEVH01020342 PNF21460.1 CH940650 EDW67337.1 CH933806 EDW16775.1 KK107323 QOIP01000002 EZA52559.1 RLU26175.1 GBXI01013550 JAD00742.1 KQ414940 KOC59235.1 KQ971372 EFA10266.1 ABLF02036082 GL442221 EFN63740.1 KZ288185 PBC34929.1 GGMR01007898 MBY20517.1 KQ982815 KYQ50216.1 KQ435180 KZC14971.1 KQ979685 KYN19774.1 KQ981606 KYN39488.1 ADTU01000542 KQ976423 KYM88890.1 GL762231 EFZ21778.1 GL888633 EGI58728.1 GL445463 EFN89546.1 GECU01005462 JAT02245.1 APGK01024354 APGK01024355 KB740544 KB632130 ENN80410.1 ERL88961.1 GGMS01004011 MBY73214.1 KQ977671 KYN00619.1 KC282411 AGX25168.1 KK852806 KDR16096.1 DS232261 EDS38649.1 PYGN01000005 PSN58281.1 AK417411 BAN20626.1 GEZM01062872 JAV69446.1 GDHF01021740 JAI30574.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000183832 UP000008820 UP000092444 UP000069940 UP000249989 UP000037069 UP000095301 UP000078200 UP000095300 UP000092460 UP000000803 UP000075881 UP000075840 UP000075882 UP000007062 UP000076407 UP000075902 UP000075903 UP000192221 UP000075885 UP000075900 UP000268350 UP000001819 UP000091820 UP000002358 UP000075901 UP000075883 UP000076408 UP000007801 UP000075884 UP000008711 UP000075886 UP000075880 UP000092443 UP000092553 UP000002282 UP000036403 UP000000304 UP000008744 UP000000673 UP000007798 UP000030765 UP000001070 UP000235965 UP000069272 UP000008792 UP000009192 UP000053097 UP000279307 UP000075920 UP000053825 UP000005203 UP000007266 UP000007819 UP000000311 UP000242457 UP000075809 UP000076502 UP000078492 UP000078541 UP000005205 UP000078540 UP000007755 UP000008237 UP000019118 UP000030742 UP000078542 UP000092445 UP000027135 UP000192223 UP000002320 UP000245037
UP000183832 UP000008820 UP000092444 UP000069940 UP000249989 UP000037069 UP000095301 UP000078200 UP000095300 UP000092460 UP000000803 UP000075881 UP000075840 UP000075882 UP000007062 UP000076407 UP000075902 UP000075903 UP000192221 UP000075885 UP000075900 UP000268350 UP000001819 UP000091820 UP000002358 UP000075901 UP000075883 UP000076408 UP000007801 UP000075884 UP000008711 UP000075886 UP000075880 UP000092443 UP000092553 UP000002282 UP000036403 UP000000304 UP000008744 UP000000673 UP000007798 UP000030765 UP000001070 UP000235965 UP000069272 UP000008792 UP000009192 UP000053097 UP000279307 UP000075920 UP000053825 UP000005203 UP000007266 UP000007819 UP000000311 UP000242457 UP000075809 UP000076502 UP000078492 UP000078541 UP000005205 UP000078540 UP000007755 UP000008237 UP000019118 UP000030742 UP000078542 UP000092445 UP000027135 UP000192223 UP000002320 UP000245037
Pfam
PF00415 RCC1
SUPFAM
SSF50985
SSF50985
Gene 3D
ProteinModelPortal
H9IS05
A0A2H1VAF6
A0A2A4JEN5
A0A2W1BC70
I4DMH0
A0A0N1INB0
+ More
A0A212FBG2 A0A194QDV3 A0A0L7KVA7 A0A1J1J651 A0A1S4F843 Q17CK1 A0A1B0G226 A0A182GGR3 A0A0L0BRC1 A0A336KMX4 A0A1I8MVK0 V5GIN9 A0A1A9VDQ1 A0A1I8QCX6 A0A1B0B2Y7 Q9VHH1 A0A182KB23 A0A182HYP1 A0A182KZI9 Q7QA05 A0A182XII8 A0A182TWG5 A0A182VA19 A0A0K8WC62 A0A1W4VQR3 A0A182PP77 A0A182RE42 A0A3B0JDK2 Q29CF9 A0A1A9W5N9 K7IPI4 A0A034VUM8 A0A182SQ60 A0A182MMQ6 A0A182Y8S4 A0A0K8UB59 B3M1L9 A0A182NGF6 B3P1T0 A0A182Q8G0 A0A182IP29 A0A1A9Y0J1 A0A0M4EQR6 B4PU09 A0A0J7NXA9 B4QX91 B4GN40 W5J9J2 B4NG19 A0A084WP51 B4JRK2 A0A2J7PYT1 A0A182F6M5 B4M0P5 B4KAN8 A0A026WBX3 A0A0A1WPG8 A0A182W3F9 A0A0L7QKX4 A0A088AFD1 D6X2F8 J9K5M4 E2AS40 A0A2A3EUV6 A0A2S2NTR6 A0A151WQS4 A0A154PST5 A0A195E4I8 A0A195FGG5 A0A158NXZ2 A0A195BS98 E9ICC8 F4X4D2 E2B432 A0A1B6JSR3 N6UF93 A0A2S2Q674 A0A151IGQ5 V9PB56 A0A1A9ZSW4 A0A067R2A3 A0A1W4X0J0 B0X1K0 A0A2P8ZP51 R4WP12 A0A1Y1LD01 A0A0K8UV50
A0A212FBG2 A0A194QDV3 A0A0L7KVA7 A0A1J1J651 A0A1S4F843 Q17CK1 A0A1B0G226 A0A182GGR3 A0A0L0BRC1 A0A336KMX4 A0A1I8MVK0 V5GIN9 A0A1A9VDQ1 A0A1I8QCX6 A0A1B0B2Y7 Q9VHH1 A0A182KB23 A0A182HYP1 A0A182KZI9 Q7QA05 A0A182XII8 A0A182TWG5 A0A182VA19 A0A0K8WC62 A0A1W4VQR3 A0A182PP77 A0A182RE42 A0A3B0JDK2 Q29CF9 A0A1A9W5N9 K7IPI4 A0A034VUM8 A0A182SQ60 A0A182MMQ6 A0A182Y8S4 A0A0K8UB59 B3M1L9 A0A182NGF6 B3P1T0 A0A182Q8G0 A0A182IP29 A0A1A9Y0J1 A0A0M4EQR6 B4PU09 A0A0J7NXA9 B4QX91 B4GN40 W5J9J2 B4NG19 A0A084WP51 B4JRK2 A0A2J7PYT1 A0A182F6M5 B4M0P5 B4KAN8 A0A026WBX3 A0A0A1WPG8 A0A182W3F9 A0A0L7QKX4 A0A088AFD1 D6X2F8 J9K5M4 E2AS40 A0A2A3EUV6 A0A2S2NTR6 A0A151WQS4 A0A154PST5 A0A195E4I8 A0A195FGG5 A0A158NXZ2 A0A195BS98 E9ICC8 F4X4D2 E2B432 A0A1B6JSR3 N6UF93 A0A2S2Q674 A0A151IGQ5 V9PB56 A0A1A9ZSW4 A0A067R2A3 A0A1W4X0J0 B0X1K0 A0A2P8ZP51 R4WP12 A0A1Y1LD01 A0A0K8UV50
Ontologies
PANTHER
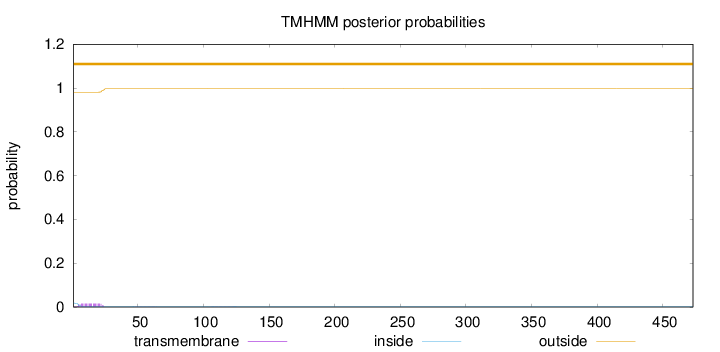
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.994683
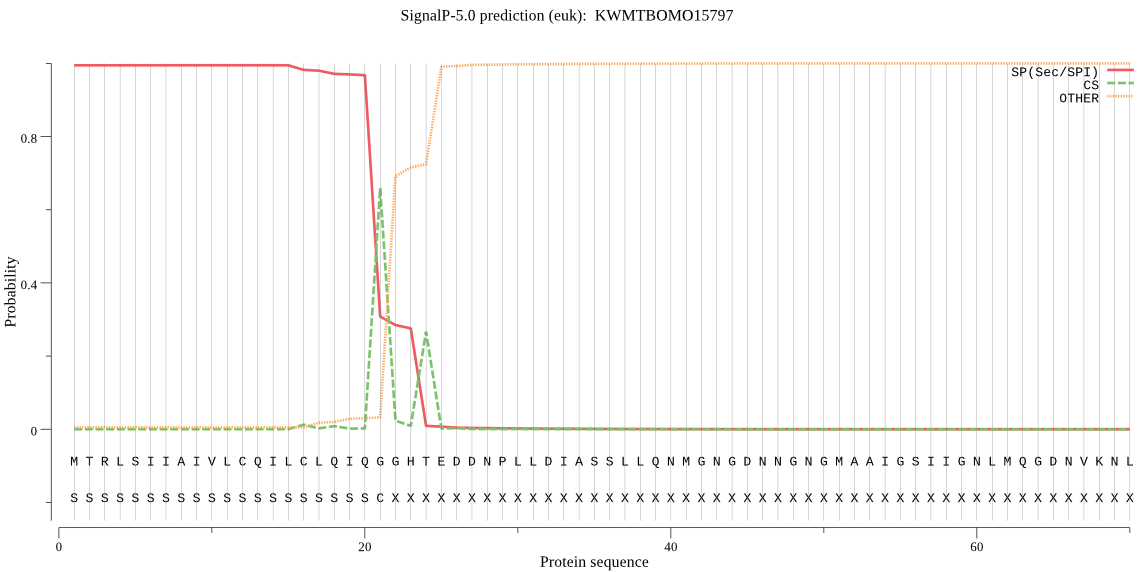
Length:
473
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.318370000000001
Exp number, first 60 AAs:
0.30172
Total prob of N-in:
0.01908
outside
1 - 473
Population Genetic Test Statistics
Pi
173.020846
Theta
21.369124
Tajima's D
-1.906638
CLR
0
CSRT
0.0186990650467477
Interpretation
Uncertain
