Gene
KWMTBOMO15795 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000029
Annotation
nucleolar_protein_family_A_member_2_[Bombyx_mori]
Full name
H/ACA ribonucleoprotein complex subunit 2-like protein
Alternative Name
H/ACA snoRNP protein NHP2
Nucleolar protein family A member 2-like protein
Nucleolar protein family A member 2-like protein
Location in the cell
Cytoplasmic Reliability : 2.486
Sequence
CDS
ATGGGTAAAGTAAAGCAAGAACCGGTCGAACAGGAAGACCAGGCTGATGTCAGCATTAAAGCTGAACCGCAAAGCCACGATGAAAAAGTCGAACATTGTAGTATAATAGCGAAACCGATGGCCACAAAAAAACTCACCAAAAAGATATATAAACTTATTAAGAAATCGAGTAACCATAAGAACTATATCAGAAACGGCCTGAAAATCGTTCAGAAGCAGTTACGTCTGGGTGAAAAGGGCATGGTTTTCTTTGCTGGAGACATTTCACCCATAGAAATCATGTGTCACCTACCTGCGGTTTGCGAGGAGAAGGACGTCCAATACTGTTACACTCCAAGTAGGAAGGATATCGGTGCCGCCATGGGTACAATGCGAGGATGCATCATGGTACTTGTCAAAGAACATGAGGATTATAAAGATTTGTACGAGGAAGTTAAAAGTGAAATAAAATTACTCGGACATCCACTGATCTGA
Protein
MGKVKQEPVEQEDQADVSIKAEPQSHDEKVEHCSIIAKPMATKKLTKKIYKLIKKSSNHKNYIRNGLKIVQKQLRLGEKGMVFFAGDISPIEIMCHLPAVCEEKDVQYCYTPSRKDIGAAMGTMRGCIMVLVKEHEDYKDLYEEVKSEIKLLGHPLI
Summary
Description
Required for ribosome biogenesis. Part of a complex which catalyzes pseudouridylation of rRNA. This involves the isomerization of uridine such that the ribose is subsequently attached to C5, instead of the normal N1. Pseudouridine ("psi") residues may serve to stabilize the conformation of rRNAs (By similarity).
Subunit
Component of the small nucleolar ribonucleoprotein particle containing H/ACA-type snoRNAs (H/ACA snoRNPs).
Similarity
Belongs to the eukaryotic ribosomal protein eL8 family.
Keywords
Complete proteome
Nucleus
Phosphoprotein
Reference proteome
Ribonucleoprotein
Ribosome biogenesis
RNA-binding
rRNA processing
Feature
chain H/ACA ribonucleoprotein complex subunit 2-like protein
Uniprot
Q1HPV4
A0A212FBF5
A0A3S2PIY0
A0A2A4JCS7
A0A2W1BF20
S4P2J6
+ More
A0A2H1VAY1 A0A0N1IFJ1 A0A194QE04 A0A1E1WNR5 A0A182N2M1 D6X2E6 A0A2M4AMH9 A0A182SQ14 A0A182RQV5 A0A182Y210 A0A182JXP8 A0A2M3ZHQ9 W5JKR9 A0A182FW52 A0A2A3EEF8 A0A088ADB0 A0A182WZA1 A0A182V2J6 A0A182HPV4 Q7Q018 A0A0L0C3X9 A0A182MD06 T1PGT4 Q17FC3 A0A182JBA6 A0A1L8EH37 T1DIY6 A0A0K8TR01 A0A1A9Z1Q9 A0A182TQ25 A0A1A9VMQ7 A0A1A9Y9U8 A0A023EFR1 A0A1B0BWM5 A0A182QYA3 A0A1A9WCS9 B0WDU0 A0A0J7KZV7 A0A182W9D3 A0A1B0FA18 A0A1I8PBN7 A0A336L7X8 A0A026WI95 T1H5M0 E2AUQ3 A0A1Q3F152 W8CAD7 A0A084WHG5 A0A1W4XP98 A0A151WED0 A0A0J9UL49 B4HHT8 M9PCI2 Q9V3U2 A0A0C9R4J3 A0A158NRF7 A0A195BDR6 A0A195FGM8 A0A0H5APS5 A0A0H5AKI8 A0A0M4EDT3 A0A195EEW6 E9IAH3 A0A0H5AKG9 A0A0H5AKJ9 A0A0H5AKH8 A0A0H5ANG0 B4LFM7 A0A1W4U8Z2 A0A151IDQ4 A0A0H5AKJ2 A0A0N8C1G9 C1BUW6 A0A0P5CK34 A0A0H5APT4 A0A0H5AKJ5 A0A310SE07 Q6XIP0 A0A0N8DJB6 A0A0L7RJK3 A0A232FCX2 B4IY17 A0A034WNU4 A0A0K8WEC2 B3MAV5 B3NHJ1 A0A1Y1MBS3 E9H7K6 A0A0P5FIY4 K7IY67 A0A3F2YUW2 B4NM18 B4KXN7 A0A3B0J7D8
A0A2H1VAY1 A0A0N1IFJ1 A0A194QE04 A0A1E1WNR5 A0A182N2M1 D6X2E6 A0A2M4AMH9 A0A182SQ14 A0A182RQV5 A0A182Y210 A0A182JXP8 A0A2M3ZHQ9 W5JKR9 A0A182FW52 A0A2A3EEF8 A0A088ADB0 A0A182WZA1 A0A182V2J6 A0A182HPV4 Q7Q018 A0A0L0C3X9 A0A182MD06 T1PGT4 Q17FC3 A0A182JBA6 A0A1L8EH37 T1DIY6 A0A0K8TR01 A0A1A9Z1Q9 A0A182TQ25 A0A1A9VMQ7 A0A1A9Y9U8 A0A023EFR1 A0A1B0BWM5 A0A182QYA3 A0A1A9WCS9 B0WDU0 A0A0J7KZV7 A0A182W9D3 A0A1B0FA18 A0A1I8PBN7 A0A336L7X8 A0A026WI95 T1H5M0 E2AUQ3 A0A1Q3F152 W8CAD7 A0A084WHG5 A0A1W4XP98 A0A151WED0 A0A0J9UL49 B4HHT8 M9PCI2 Q9V3U2 A0A0C9R4J3 A0A158NRF7 A0A195BDR6 A0A195FGM8 A0A0H5APS5 A0A0H5AKI8 A0A0M4EDT3 A0A195EEW6 E9IAH3 A0A0H5AKG9 A0A0H5AKJ9 A0A0H5AKH8 A0A0H5ANG0 B4LFM7 A0A1W4U8Z2 A0A151IDQ4 A0A0H5AKJ2 A0A0N8C1G9 C1BUW6 A0A0P5CK34 A0A0H5APT4 A0A0H5AKJ5 A0A310SE07 Q6XIP0 A0A0N8DJB6 A0A0L7RJK3 A0A232FCX2 B4IY17 A0A034WNU4 A0A0K8WEC2 B3MAV5 B3NHJ1 A0A1Y1MBS3 E9H7K6 A0A0P5FIY4 K7IY67 A0A3F2YUW2 B4NM18 B4KXN7 A0A3B0J7D8
Pubmed
19121390
22118469
28756777
23622113
26354079
18362917
+ More
19820115 25244985 20920257 23761445 12364791 14747013 17210077 26108605 25315136 17510324 24330624 26369729 24945155 26483478 24508170 30249741 20798317 24495485 24438588 22936249 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18327897 21347285 21282665 14525923 28648823 25348373 28004739 21292972 20075255 20966253
19820115 25244985 20920257 23761445 12364791 14747013 17210077 26108605 25315136 17510324 24330624 26369729 24945155 26483478 24508170 30249741 20798317 24495485 24438588 22936249 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18327897 21347285 21282665 14525923 28648823 25348373 28004739 21292972 20075255 20966253
EMBL
BABH01035561
DQ443298
ABF51387.1
AGBW02009344
OWR51060.1
RSAL01000019
+ More
RVE52745.1 NWSH01001860 PCG69885.1 KZ150146 PZC72891.1 GAIX01011980 JAA80580.1 ODYU01001334 SOQ37374.1 KQ460273 KPJ16327.1 KQ459144 KPJ03654.1 GDQN01002573 JAT88481.1 KQ971371 EFA09871.1 GGFK01008663 MBW41984.1 GGFM01007343 MBW28094.1 ADMH02001151 ETN63893.1 KZ288271 PBC29854.1 APCN01003277 AAAB01008986 EAA00120.2 JRES01000945 KNC26931.1 AXCM01006131 KA647966 AFP62595.1 CH477272 EAT45222.1 GFDG01000825 JAV17974.1 GALA01000661 JAA94191.1 GDAI01000811 JAI16792.1 JXUM01015996 JXUM01042476 GAPW01005416 KQ561326 KQ560445 JAC08182.1 KXJ78975.1 KXJ82382.1 JXJN01021871 AXCN02001078 DS231901 EDS44877.1 LBMM01001707 KMQ95863.1 CCAG010018114 UFQS01001854 UFQT01001854 SSX12575.1 SSX32018.1 KK107193 QOIP01000002 EZA55683.1 RLU25599.1 GL442872 EFN62852.1 GFDL01013760 JAV21285.1 GAMC01001394 JAC05162.1 ATLV01023814 KE525347 KFB49659.1 KQ983238 KYQ46223.1 CM002912 KMY99630.1 CH480815 EDW41503.1 AE014296 KX531559 AGB94542.1 ANY27369.1 AF217228 BT023752 GBYB01002840 JAG72607.1 ADTU01000402 KQ976509 KYM82711.1 KQ981606 KYN39548.1 LC058863 BAR89708.1 LC058844 LC058849 LC058852 LC058858 BAR89689.1 BAR89694.1 BAR89697.1 BAR89703.1 CP012525 ALC43808.1 KQ979039 KYN23347.1 GL762022 EFZ22450.1 LC058830 LC058831 LC058832 LC058833 LC058835 LC058836 LC058837 LC058838 LC058840 LC058841 LC058842 LC058843 LC058845 LC058847 LC058848 LC058851 LC058855 LC058856 LC058857 LC058860 LC058861 BAR89675.1 BAR89676.1 BAR89677.1 BAR89678.1 BAR89680.1 BAR89681.1 BAR89682.1 BAR89683.1 BAR89685.1 BAR89686.1 BAR89687.1 BAR89688.1 BAR89690.1 BAR89700.1 BAR89705.1 LC058864 LC058865 LC058867 LC058868 LC058870 LC058874 LC058875 LC058876 LC058877 BAR89709.1 BAR89710.1 BAR89712.1 BAR89713.1 BAR89715.1 BAR89719.1 BAR89720.1 BAR89721.1 BAR89722.1 LC058834 LC058853 BAR89679.1 BAR89698.1 LC058862 LC058866 LC058869 LC058871 LC058872 BAR89707.1 BAR89711.1 BAR89714.1 BAR89716.1 BAR89717.1 CH940647 EDW70345.1 KQ977978 KYM98379.1 LC058846 LC058854 BAR89691.1 BAR89699.1 GDIQ01124107 LRGB01000311 JAL27619.1 KZS19787.1 BT078395 BT120685 ACO12819.1 ADD24325.1 GDIP01185056 JAJ38346.1 LC058873 BAR89718.1 LC058839 LC058850 LC058859 BAR89684.1 BAR89695.1 BAR89704.1 KQ760827 OAD58898.1 AY231789 CM000159 GDIP01028119 JAM75596.1 KQ414581 KOC70978.1 NNAY01000399 OXU28654.1 CH916366 EDV97560.1 GAKP01003509 JAC55443.1 GDHF01027221 GDHF01002872 JAI25093.1 JAI49442.1 CH902618 EDV39189.1 CH954178 EDV51786.1 GEZM01037624 JAV82020.1 GL732601 EFX72202.1 GDIQ01256438 JAJ95286.1 CH964278 EDW85386.1 CH933809 EDW19744.1 OUUW01000002 SPP76043.1
RVE52745.1 NWSH01001860 PCG69885.1 KZ150146 PZC72891.1 GAIX01011980 JAA80580.1 ODYU01001334 SOQ37374.1 KQ460273 KPJ16327.1 KQ459144 KPJ03654.1 GDQN01002573 JAT88481.1 KQ971371 EFA09871.1 GGFK01008663 MBW41984.1 GGFM01007343 MBW28094.1 ADMH02001151 ETN63893.1 KZ288271 PBC29854.1 APCN01003277 AAAB01008986 EAA00120.2 JRES01000945 KNC26931.1 AXCM01006131 KA647966 AFP62595.1 CH477272 EAT45222.1 GFDG01000825 JAV17974.1 GALA01000661 JAA94191.1 GDAI01000811 JAI16792.1 JXUM01015996 JXUM01042476 GAPW01005416 KQ561326 KQ560445 JAC08182.1 KXJ78975.1 KXJ82382.1 JXJN01021871 AXCN02001078 DS231901 EDS44877.1 LBMM01001707 KMQ95863.1 CCAG010018114 UFQS01001854 UFQT01001854 SSX12575.1 SSX32018.1 KK107193 QOIP01000002 EZA55683.1 RLU25599.1 GL442872 EFN62852.1 GFDL01013760 JAV21285.1 GAMC01001394 JAC05162.1 ATLV01023814 KE525347 KFB49659.1 KQ983238 KYQ46223.1 CM002912 KMY99630.1 CH480815 EDW41503.1 AE014296 KX531559 AGB94542.1 ANY27369.1 AF217228 BT023752 GBYB01002840 JAG72607.1 ADTU01000402 KQ976509 KYM82711.1 KQ981606 KYN39548.1 LC058863 BAR89708.1 LC058844 LC058849 LC058852 LC058858 BAR89689.1 BAR89694.1 BAR89697.1 BAR89703.1 CP012525 ALC43808.1 KQ979039 KYN23347.1 GL762022 EFZ22450.1 LC058830 LC058831 LC058832 LC058833 LC058835 LC058836 LC058837 LC058838 LC058840 LC058841 LC058842 LC058843 LC058845 LC058847 LC058848 LC058851 LC058855 LC058856 LC058857 LC058860 LC058861 BAR89675.1 BAR89676.1 BAR89677.1 BAR89678.1 BAR89680.1 BAR89681.1 BAR89682.1 BAR89683.1 BAR89685.1 BAR89686.1 BAR89687.1 BAR89688.1 BAR89690.1 BAR89700.1 BAR89705.1 LC058864 LC058865 LC058867 LC058868 LC058870 LC058874 LC058875 LC058876 LC058877 BAR89709.1 BAR89710.1 BAR89712.1 BAR89713.1 BAR89715.1 BAR89719.1 BAR89720.1 BAR89721.1 BAR89722.1 LC058834 LC058853 BAR89679.1 BAR89698.1 LC058862 LC058866 LC058869 LC058871 LC058872 BAR89707.1 BAR89711.1 BAR89714.1 BAR89716.1 BAR89717.1 CH940647 EDW70345.1 KQ977978 KYM98379.1 LC058846 LC058854 BAR89691.1 BAR89699.1 GDIQ01124107 LRGB01000311 JAL27619.1 KZS19787.1 BT078395 BT120685 ACO12819.1 ADD24325.1 GDIP01185056 JAJ38346.1 LC058873 BAR89718.1 LC058839 LC058850 LC058859 BAR89684.1 BAR89695.1 BAR89704.1 KQ760827 OAD58898.1 AY231789 CM000159 GDIP01028119 JAM75596.1 KQ414581 KOC70978.1 NNAY01000399 OXU28654.1 CH916366 EDV97560.1 GAKP01003509 JAC55443.1 GDHF01027221 GDHF01002872 JAI25093.1 JAI49442.1 CH902618 EDV39189.1 CH954178 EDV51786.1 GEZM01037624 JAV82020.1 GL732601 EFX72202.1 GDIQ01256438 JAJ95286.1 CH964278 EDW85386.1 CH933809 EDW19744.1 OUUW01000002 SPP76043.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000218220
UP000053240
UP000053268
+ More
UP000075884 UP000007266 UP000075901 UP000075900 UP000076408 UP000075881 UP000000673 UP000069272 UP000242457 UP000005203 UP000076407 UP000075903 UP000075840 UP000007062 UP000037069 UP000075883 UP000095301 UP000008820 UP000075880 UP000092445 UP000075902 UP000078200 UP000092443 UP000069940 UP000249989 UP000092460 UP000075886 UP000091820 UP000002320 UP000036403 UP000075920 UP000092444 UP000095300 UP000053097 UP000279307 UP000015102 UP000000311 UP000030765 UP000192223 UP000075809 UP000001292 UP000000803 UP000005205 UP000078540 UP000078541 UP000092553 UP000078492 UP000008792 UP000192221 UP000078542 UP000076858 UP000002282 UP000053825 UP000215335 UP000001070 UP000007801 UP000008711 UP000000305 UP000002358 UP000075882 UP000007798 UP000009192 UP000268350
UP000075884 UP000007266 UP000075901 UP000075900 UP000076408 UP000075881 UP000000673 UP000069272 UP000242457 UP000005203 UP000076407 UP000075903 UP000075840 UP000007062 UP000037069 UP000075883 UP000095301 UP000008820 UP000075880 UP000092445 UP000075902 UP000078200 UP000092443 UP000069940 UP000249989 UP000092460 UP000075886 UP000091820 UP000002320 UP000036403 UP000075920 UP000092444 UP000095300 UP000053097 UP000279307 UP000015102 UP000000311 UP000030765 UP000192223 UP000075809 UP000001292 UP000000803 UP000005205 UP000078540 UP000078541 UP000092553 UP000078492 UP000008792 UP000192221 UP000078542 UP000076858 UP000002282 UP000053825 UP000215335 UP000001070 UP000007801 UP000008711 UP000000305 UP000002358 UP000075882 UP000007798 UP000009192 UP000268350
Pfam
PF01248 Ribosomal_L7Ae
Interpro
SUPFAM
SSF55315
SSF55315
Gene 3D
ProteinModelPortal
Q1HPV4
A0A212FBF5
A0A3S2PIY0
A0A2A4JCS7
A0A2W1BF20
S4P2J6
+ More
A0A2H1VAY1 A0A0N1IFJ1 A0A194QE04 A0A1E1WNR5 A0A182N2M1 D6X2E6 A0A2M4AMH9 A0A182SQ14 A0A182RQV5 A0A182Y210 A0A182JXP8 A0A2M3ZHQ9 W5JKR9 A0A182FW52 A0A2A3EEF8 A0A088ADB0 A0A182WZA1 A0A182V2J6 A0A182HPV4 Q7Q018 A0A0L0C3X9 A0A182MD06 T1PGT4 Q17FC3 A0A182JBA6 A0A1L8EH37 T1DIY6 A0A0K8TR01 A0A1A9Z1Q9 A0A182TQ25 A0A1A9VMQ7 A0A1A9Y9U8 A0A023EFR1 A0A1B0BWM5 A0A182QYA3 A0A1A9WCS9 B0WDU0 A0A0J7KZV7 A0A182W9D3 A0A1B0FA18 A0A1I8PBN7 A0A336L7X8 A0A026WI95 T1H5M0 E2AUQ3 A0A1Q3F152 W8CAD7 A0A084WHG5 A0A1W4XP98 A0A151WED0 A0A0J9UL49 B4HHT8 M9PCI2 Q9V3U2 A0A0C9R4J3 A0A158NRF7 A0A195BDR6 A0A195FGM8 A0A0H5APS5 A0A0H5AKI8 A0A0M4EDT3 A0A195EEW6 E9IAH3 A0A0H5AKG9 A0A0H5AKJ9 A0A0H5AKH8 A0A0H5ANG0 B4LFM7 A0A1W4U8Z2 A0A151IDQ4 A0A0H5AKJ2 A0A0N8C1G9 C1BUW6 A0A0P5CK34 A0A0H5APT4 A0A0H5AKJ5 A0A310SE07 Q6XIP0 A0A0N8DJB6 A0A0L7RJK3 A0A232FCX2 B4IY17 A0A034WNU4 A0A0K8WEC2 B3MAV5 B3NHJ1 A0A1Y1MBS3 E9H7K6 A0A0P5FIY4 K7IY67 A0A3F2YUW2 B4NM18 B4KXN7 A0A3B0J7D8
A0A2H1VAY1 A0A0N1IFJ1 A0A194QE04 A0A1E1WNR5 A0A182N2M1 D6X2E6 A0A2M4AMH9 A0A182SQ14 A0A182RQV5 A0A182Y210 A0A182JXP8 A0A2M3ZHQ9 W5JKR9 A0A182FW52 A0A2A3EEF8 A0A088ADB0 A0A182WZA1 A0A182V2J6 A0A182HPV4 Q7Q018 A0A0L0C3X9 A0A182MD06 T1PGT4 Q17FC3 A0A182JBA6 A0A1L8EH37 T1DIY6 A0A0K8TR01 A0A1A9Z1Q9 A0A182TQ25 A0A1A9VMQ7 A0A1A9Y9U8 A0A023EFR1 A0A1B0BWM5 A0A182QYA3 A0A1A9WCS9 B0WDU0 A0A0J7KZV7 A0A182W9D3 A0A1B0FA18 A0A1I8PBN7 A0A336L7X8 A0A026WI95 T1H5M0 E2AUQ3 A0A1Q3F152 W8CAD7 A0A084WHG5 A0A1W4XP98 A0A151WED0 A0A0J9UL49 B4HHT8 M9PCI2 Q9V3U2 A0A0C9R4J3 A0A158NRF7 A0A195BDR6 A0A195FGM8 A0A0H5APS5 A0A0H5AKI8 A0A0M4EDT3 A0A195EEW6 E9IAH3 A0A0H5AKG9 A0A0H5AKJ9 A0A0H5AKH8 A0A0H5ANG0 B4LFM7 A0A1W4U8Z2 A0A151IDQ4 A0A0H5AKJ2 A0A0N8C1G9 C1BUW6 A0A0P5CK34 A0A0H5APT4 A0A0H5AKJ5 A0A310SE07 Q6XIP0 A0A0N8DJB6 A0A0L7RJK3 A0A232FCX2 B4IY17 A0A034WNU4 A0A0K8WEC2 B3MAV5 B3NHJ1 A0A1Y1MBS3 E9H7K6 A0A0P5FIY4 K7IY67 A0A3F2YUW2 B4NM18 B4KXN7 A0A3B0J7D8
PDB
2LBX
E-value=1.52971e-13,
Score=179
Ontologies
GO
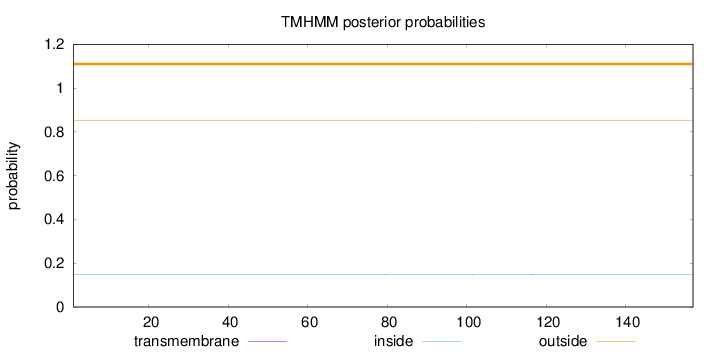
Topology
Subcellular location
Length:
157
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01357
Exp number, first 60 AAs:
0
Total prob of N-in:
0.14932
outside
1 - 157
Population Genetic Test Statistics
Pi
103.359744
Theta
110.445381
Tajima's D
-0.206554
CLR
1.494316
CSRT
0.309434528273586
Interpretation
Uncertain
