Gene
KWMTBOMO15793
Annotation
PREDICTED:_rho-related_BTB_domain-containing_protein_1-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.419
Sequence
CDS
ATGGACAACGAGCAACCGCATCAAGAACTGGTGAAATGCGTGGTGGTCGGAGACACGGCCGTGGGGAAGACGCGCTTAATATGCGCCAGGGCTTGCAACAAACACGTTTCACTGTCGCAGTTAATGACCACTCATGTCCCTACCGTGTGGGCCATTGACCAGTACAGGATTTACAAGGACGTATTAGAAAGATCCTGGGAAATGGTCGACGGAGTGAACGTGTCGTTGCGTTTGTGGGACACATTCGGCGATCACGAAAAGGACAGGAGATTTGCGTATGGAAGATCCGATGTTGTCCTGCTGTGTTTTTCAATAACGAATCCTATATCGTTGAGAAACTGTGGAGCTATGTGGTATCCAGAAATCAGACGTTTTTGCCCGAACACTCCGATCCTGCTAGTCGGTTGCAAGAACGATTTGCGATACATGTACAGAGACGAGAATTACCTTAGCTACTGCAAGGACCGGAGCCCTTTCATAAGGGCTCCAAGGAAATGCGACTTGGTCATGCCGGACCAAGGTCGAGCCTTAGCTCATGAGTTCGGCATTTATTATTACGAAACCTCAGTGTTCACATACTACGGAGTGAACGAAGTCTTCGAGAATGCCATCAGGGCGGCGCTGATGGCGAGGAGACAACAGAGGTTCTGGATGACCAATTTAAAGAGAGTTCAGCGGCCTCTTCAGCAGGCGCCGTTCAGACCACCTAGACCAGTGGAGCCCGAAGTCACTTTCGTCAGCAGTAATTACCTCGAGAATATGACTAATTTACTCCATCAGCAGGTAATTTATTAA
Protein
MDNEQPHQELVKCVVVGDTAVGKTRLICARACNKHVSLSQLMTTHVPTVWAIDQYRIYKDVLERSWEMVDGVNVSLRLWDTFGDHEKDRRFAYGRSDVVLLCFSITNPISLRNCGAMWYPEIRRFCPNTPILLVGCKNDLRYMYRDENYLSYCKDRSPFIRAPRKCDLVMPDQGRALAHEFGIYYYETSVFTYYGVNEVFENAIRAALMARRQQRFWMTNLKRVQRPLQQAPFRPPRPVEPEVTFVSSNYLENMTNLLHQQVIY
Summary
Uniprot
A0A2W1BC97
A0A2A4JDM4
A0A3S2NJR1
A0A0N0PAL4
A0A212FBE1
A0A194QFD4
+ More
A0A2H1WD79 A0A2J7PCZ5 A0A2J7PD07 A0A0C9S165 A0A1B6I3T6 A0A2P8XD96 V9IB50 A0A0J7L8D1 A0A088ADH9 A0A310SIA9 E9IAH5 A0A0C9RYS7 E2BJW9 A0A0C9S162 A0A154P1W4 F4WJL0 A0A158NRQ5 A0A0L7RJG7 A0A151WEK8 A0A151IDA7 A0A0M9A9T1 A0A195FH34 A0A026WKQ9 A0A3L8DYT0 A0A1B6FT63 A0A195BDT0 K7IUB5 A0A232F667 A0A2J7PD06 A0A1B6KCB0 A0A1B6M0T0 A0A1J1IWE1 A0A336LIY4 A0A182JL30 B0WY87 A0A1Q3G4P4 A0A1Q3G4P9 A0A1Q3G4Q1 A0A1Q3G4T5 A0A1L8DML6 A0A182WC15 A0A182X921 A0A182RR36 A0A182M590 Q16FU0 Q7Q5F0 Q17F37 A0A182NJ17 A0A182H480 A0A182PPG4 A0A182I459 A0A182LKV2 U5EWH4 A0A2M4A1A5 A0A182FDY0 A0A2M3YYD2 A0A2M3YYE0 A0A2A3EDJ8 W5JG69 A0A182YFI6 A0A1B6HQC0 A0A182Q319 A0A0A9WW42 D6X2E5 A0A1A9WBP4 A0A1A9V2Z1 A0A0A1XFU8 W8BQA2 W8AS84 A0A1B0FBL0 A0A1A9ZFX7 A0A0A1X0E0 A0A1A9YL37 A0A1B0ARA9 A0A0K8UXW8 A0A0A1XTE9 A0A0K8VRY5 A0A034VMB4 A0A1I8NNF8 A0A1I8NNF6 A0A1I8MXE2 A0A1Y1M856 A0A0L0CAT0 N6U401 B3M984 A0A067QTG6 B4IA94 B4QRY1 A0A1W4W6L7 E0VA13 A0A0J9RYN2 M9PIB5 B3NIK6 A0A0J9RYV9
A0A2H1WD79 A0A2J7PCZ5 A0A2J7PD07 A0A0C9S165 A0A1B6I3T6 A0A2P8XD96 V9IB50 A0A0J7L8D1 A0A088ADH9 A0A310SIA9 E9IAH5 A0A0C9RYS7 E2BJW9 A0A0C9S162 A0A154P1W4 F4WJL0 A0A158NRQ5 A0A0L7RJG7 A0A151WEK8 A0A151IDA7 A0A0M9A9T1 A0A195FH34 A0A026WKQ9 A0A3L8DYT0 A0A1B6FT63 A0A195BDT0 K7IUB5 A0A232F667 A0A2J7PD06 A0A1B6KCB0 A0A1B6M0T0 A0A1J1IWE1 A0A336LIY4 A0A182JL30 B0WY87 A0A1Q3G4P4 A0A1Q3G4P9 A0A1Q3G4Q1 A0A1Q3G4T5 A0A1L8DML6 A0A182WC15 A0A182X921 A0A182RR36 A0A182M590 Q16FU0 Q7Q5F0 Q17F37 A0A182NJ17 A0A182H480 A0A182PPG4 A0A182I459 A0A182LKV2 U5EWH4 A0A2M4A1A5 A0A182FDY0 A0A2M3YYD2 A0A2M3YYE0 A0A2A3EDJ8 W5JG69 A0A182YFI6 A0A1B6HQC0 A0A182Q319 A0A0A9WW42 D6X2E5 A0A1A9WBP4 A0A1A9V2Z1 A0A0A1XFU8 W8BQA2 W8AS84 A0A1B0FBL0 A0A1A9ZFX7 A0A0A1X0E0 A0A1A9YL37 A0A1B0ARA9 A0A0K8UXW8 A0A0A1XTE9 A0A0K8VRY5 A0A034VMB4 A0A1I8NNF8 A0A1I8NNF6 A0A1I8MXE2 A0A1Y1M856 A0A0L0CAT0 N6U401 B3M984 A0A067QTG6 B4IA94 B4QRY1 A0A1W4W6L7 E0VA13 A0A0J9RYN2 M9PIB5 B3NIK6 A0A0J9RYV9
Pubmed
28756777
26354079
22118469
29403074
21282665
20798317
+ More
21719571 21347285 24508170 30249741 20075255 28648823 17510324 12364791 14747013 17210077 26483478 20966253 20920257 23761445 25244985 25401762 18362917 19820115 25830018 24495485 25348373 25315136 28004739 26108605 23537049 17994087 24845553 20566863 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
21719571 21347285 24508170 30249741 20075255 28648823 17510324 12364791 14747013 17210077 26483478 20966253 20920257 23761445 25244985 25401762 18362917 19820115 25830018 24495485 25348373 25315136 28004739 26108605 23537049 17994087 24845553 20566863 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
EMBL
KZ150146
PZC72892.1
NWSH01001860
PCG69886.1
RSAL01000019
RVE52746.1
+ More
KQ461188 KPJ07310.1 AGBW02009344 OWR51061.1 KQ459144 KPJ03655.1 ODYU01007874 SOQ51028.1 NEVH01026395 PNF14206.1 PNF14207.1 GBYB01014476 JAG84243.1 GECU01026110 GECU01014188 JAS81596.1 JAS93518.1 PYGN01002785 PSN29969.1 JR037040 AEY57544.1 LBMM01000261 KMR02521.1 KQ760827 OAD58899.1 GL762022 EFZ22424.1 GBYB01014475 JAG84242.1 GL448708 EFN83978.1 GBYB01014471 JAG84238.1 KQ434803 KZC05925.1 GL888184 EGI65626.1 ADTU01000403 ADTU01000404 KQ414581 KOC70979.1 KQ983238 KYQ46221.1 KQ977978 KYM98378.1 KQ435706 KOX80130.1 KQ981606 KYN39547.1 KK107193 EZA55684.1 QOIP01000002 RLU25597.1 GECZ01016566 JAS53203.1 KQ976509 KYM82713.1 AAZX01007296 NNAY01000872 OXU26125.1 PNF14208.1 GEBQ01030889 GEBQ01023174 JAT09088.1 JAT16803.1 GEBQ01010445 JAT29532.1 CVRI01000059 CRL03470.1 UFQT01000016 SSX17800.1 DS232185 EDS36964.1 GFDL01000270 JAV34775.1 GFDL01000268 JAV34777.1 GFDL01000272 JAV34773.1 GFDL01000269 JAV34776.1 GFDF01006433 JAV07651.1 AXCM01005449 CH478381 EAT33106.1 AAAB01008960 EAA11877.4 CH477277 EAT45094.1 JXUM01109046 JXUM01109047 JXUM01109048 JXUM01109049 JXUM01109050 APCN01002274 GANO01001417 JAB58454.1 GGFK01001273 MBW34594.1 GGFM01000509 MBW21260.1 GGFM01000510 MBW21261.1 KZ288271 PBC29853.1 ADMH02001489 ETN62333.1 GECU01030828 JAS76878.1 AXCN02000460 GBHO01034536 GBHO01034534 JAG09068.1 JAG09070.1 KQ971371 EFA10257.1 GBXI01004914 JAD09378.1 GAMC01014831 GAMC01014829 JAB91724.1 GAMC01014830 JAB91725.1 CCAG010010279 GBXI01009705 JAD04587.1 JXJN01002298 GDHF01020797 JAI31517.1 GBXI01000017 JAD14275.1 GDHF01010706 JAI41608.1 GAKP01016284 JAC42668.1 GEZM01037624 JAV82019.1 JRES01000760 KNC28564.1 APGK01050287 APGK01050288 APGK01050289 KB741169 KB632294 ENN73312.1 ERL91669.1 CH902618 EDV40068.2 KK853292 KDR08853.1 CH480826 EDW44207.1 CM000363 EDX11227.1 DS235004 EEB10219.1 CM002912 KMZ00767.1 AE014296 AGB94801.1 CH954178 EDV52502.1 KMZ00768.1
KQ461188 KPJ07310.1 AGBW02009344 OWR51061.1 KQ459144 KPJ03655.1 ODYU01007874 SOQ51028.1 NEVH01026395 PNF14206.1 PNF14207.1 GBYB01014476 JAG84243.1 GECU01026110 GECU01014188 JAS81596.1 JAS93518.1 PYGN01002785 PSN29969.1 JR037040 AEY57544.1 LBMM01000261 KMR02521.1 KQ760827 OAD58899.1 GL762022 EFZ22424.1 GBYB01014475 JAG84242.1 GL448708 EFN83978.1 GBYB01014471 JAG84238.1 KQ434803 KZC05925.1 GL888184 EGI65626.1 ADTU01000403 ADTU01000404 KQ414581 KOC70979.1 KQ983238 KYQ46221.1 KQ977978 KYM98378.1 KQ435706 KOX80130.1 KQ981606 KYN39547.1 KK107193 EZA55684.1 QOIP01000002 RLU25597.1 GECZ01016566 JAS53203.1 KQ976509 KYM82713.1 AAZX01007296 NNAY01000872 OXU26125.1 PNF14208.1 GEBQ01030889 GEBQ01023174 JAT09088.1 JAT16803.1 GEBQ01010445 JAT29532.1 CVRI01000059 CRL03470.1 UFQT01000016 SSX17800.1 DS232185 EDS36964.1 GFDL01000270 JAV34775.1 GFDL01000268 JAV34777.1 GFDL01000272 JAV34773.1 GFDL01000269 JAV34776.1 GFDF01006433 JAV07651.1 AXCM01005449 CH478381 EAT33106.1 AAAB01008960 EAA11877.4 CH477277 EAT45094.1 JXUM01109046 JXUM01109047 JXUM01109048 JXUM01109049 JXUM01109050 APCN01002274 GANO01001417 JAB58454.1 GGFK01001273 MBW34594.1 GGFM01000509 MBW21260.1 GGFM01000510 MBW21261.1 KZ288271 PBC29853.1 ADMH02001489 ETN62333.1 GECU01030828 JAS76878.1 AXCN02000460 GBHO01034536 GBHO01034534 JAG09068.1 JAG09070.1 KQ971371 EFA10257.1 GBXI01004914 JAD09378.1 GAMC01014831 GAMC01014829 JAB91724.1 GAMC01014830 JAB91725.1 CCAG010010279 GBXI01009705 JAD04587.1 JXJN01002298 GDHF01020797 JAI31517.1 GBXI01000017 JAD14275.1 GDHF01010706 JAI41608.1 GAKP01016284 JAC42668.1 GEZM01037624 JAV82019.1 JRES01000760 KNC28564.1 APGK01050287 APGK01050288 APGK01050289 KB741169 KB632294 ENN73312.1 ERL91669.1 CH902618 EDV40068.2 KK853292 KDR08853.1 CH480826 EDW44207.1 CM000363 EDX11227.1 DS235004 EEB10219.1 CM002912 KMZ00767.1 AE014296 AGB94801.1 CH954178 EDV52502.1 KMZ00768.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000007151
UP000053268
UP000235965
+ More
UP000245037 UP000036403 UP000005203 UP000008237 UP000076502 UP000007755 UP000005205 UP000053825 UP000075809 UP000078542 UP000053105 UP000078541 UP000053097 UP000279307 UP000078540 UP000002358 UP000215335 UP000183832 UP000075880 UP000002320 UP000075920 UP000076407 UP000075900 UP000075883 UP000008820 UP000007062 UP000075884 UP000069940 UP000075885 UP000075840 UP000075882 UP000069272 UP000242457 UP000000673 UP000076408 UP000075886 UP000007266 UP000091820 UP000078200 UP000092444 UP000092445 UP000092443 UP000092460 UP000095300 UP000095301 UP000037069 UP000019118 UP000030742 UP000007801 UP000027135 UP000001292 UP000000304 UP000192221 UP000009046 UP000000803 UP000008711
UP000245037 UP000036403 UP000005203 UP000008237 UP000076502 UP000007755 UP000005205 UP000053825 UP000075809 UP000078542 UP000053105 UP000078541 UP000053097 UP000279307 UP000078540 UP000002358 UP000215335 UP000183832 UP000075880 UP000002320 UP000075920 UP000076407 UP000075900 UP000075883 UP000008820 UP000007062 UP000075884 UP000069940 UP000075885 UP000075840 UP000075882 UP000069272 UP000242457 UP000000673 UP000076408 UP000075886 UP000007266 UP000091820 UP000078200 UP000092444 UP000092445 UP000092443 UP000092460 UP000095300 UP000095301 UP000037069 UP000019118 UP000030742 UP000007801 UP000027135 UP000001292 UP000000304 UP000192221 UP000009046 UP000000803 UP000008711
Interpro
ProteinModelPortal
A0A2W1BC97
A0A2A4JDM4
A0A3S2NJR1
A0A0N0PAL4
A0A212FBE1
A0A194QFD4
+ More
A0A2H1WD79 A0A2J7PCZ5 A0A2J7PD07 A0A0C9S165 A0A1B6I3T6 A0A2P8XD96 V9IB50 A0A0J7L8D1 A0A088ADH9 A0A310SIA9 E9IAH5 A0A0C9RYS7 E2BJW9 A0A0C9S162 A0A154P1W4 F4WJL0 A0A158NRQ5 A0A0L7RJG7 A0A151WEK8 A0A151IDA7 A0A0M9A9T1 A0A195FH34 A0A026WKQ9 A0A3L8DYT0 A0A1B6FT63 A0A195BDT0 K7IUB5 A0A232F667 A0A2J7PD06 A0A1B6KCB0 A0A1B6M0T0 A0A1J1IWE1 A0A336LIY4 A0A182JL30 B0WY87 A0A1Q3G4P4 A0A1Q3G4P9 A0A1Q3G4Q1 A0A1Q3G4T5 A0A1L8DML6 A0A182WC15 A0A182X921 A0A182RR36 A0A182M590 Q16FU0 Q7Q5F0 Q17F37 A0A182NJ17 A0A182H480 A0A182PPG4 A0A182I459 A0A182LKV2 U5EWH4 A0A2M4A1A5 A0A182FDY0 A0A2M3YYD2 A0A2M3YYE0 A0A2A3EDJ8 W5JG69 A0A182YFI6 A0A1B6HQC0 A0A182Q319 A0A0A9WW42 D6X2E5 A0A1A9WBP4 A0A1A9V2Z1 A0A0A1XFU8 W8BQA2 W8AS84 A0A1B0FBL0 A0A1A9ZFX7 A0A0A1X0E0 A0A1A9YL37 A0A1B0ARA9 A0A0K8UXW8 A0A0A1XTE9 A0A0K8VRY5 A0A034VMB4 A0A1I8NNF8 A0A1I8NNF6 A0A1I8MXE2 A0A1Y1M856 A0A0L0CAT0 N6U401 B3M984 A0A067QTG6 B4IA94 B4QRY1 A0A1W4W6L7 E0VA13 A0A0J9RYN2 M9PIB5 B3NIK6 A0A0J9RYV9
A0A2H1WD79 A0A2J7PCZ5 A0A2J7PD07 A0A0C9S165 A0A1B6I3T6 A0A2P8XD96 V9IB50 A0A0J7L8D1 A0A088ADH9 A0A310SIA9 E9IAH5 A0A0C9RYS7 E2BJW9 A0A0C9S162 A0A154P1W4 F4WJL0 A0A158NRQ5 A0A0L7RJG7 A0A151WEK8 A0A151IDA7 A0A0M9A9T1 A0A195FH34 A0A026WKQ9 A0A3L8DYT0 A0A1B6FT63 A0A195BDT0 K7IUB5 A0A232F667 A0A2J7PD06 A0A1B6KCB0 A0A1B6M0T0 A0A1J1IWE1 A0A336LIY4 A0A182JL30 B0WY87 A0A1Q3G4P4 A0A1Q3G4P9 A0A1Q3G4Q1 A0A1Q3G4T5 A0A1L8DML6 A0A182WC15 A0A182X921 A0A182RR36 A0A182M590 Q16FU0 Q7Q5F0 Q17F37 A0A182NJ17 A0A182H480 A0A182PPG4 A0A182I459 A0A182LKV2 U5EWH4 A0A2M4A1A5 A0A182FDY0 A0A2M3YYD2 A0A2M3YYE0 A0A2A3EDJ8 W5JG69 A0A182YFI6 A0A1B6HQC0 A0A182Q319 A0A0A9WW42 D6X2E5 A0A1A9WBP4 A0A1A9V2Z1 A0A0A1XFU8 W8BQA2 W8AS84 A0A1B0FBL0 A0A1A9ZFX7 A0A0A1X0E0 A0A1A9YL37 A0A1B0ARA9 A0A0K8UXW8 A0A0A1XTE9 A0A0K8VRY5 A0A034VMB4 A0A1I8NNF8 A0A1I8NNF6 A0A1I8MXE2 A0A1Y1M856 A0A0L0CAT0 N6U401 B3M984 A0A067QTG6 B4IA94 B4QRY1 A0A1W4W6L7 E0VA13 A0A0J9RYN2 M9PIB5 B3NIK6 A0A0J9RYV9
PDB
3RYT
E-value=4.40662e-32,
Score=342
Ontologies
GO
Topology
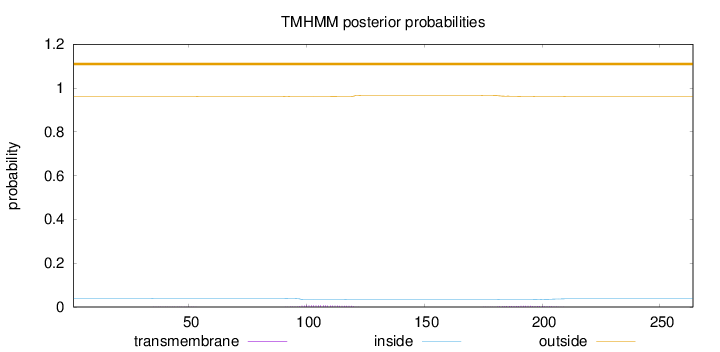
Length:
264
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.26111
Exp number, first 60 AAs:
0.007
Total prob of N-in:
0.03937
outside
1 - 264
Population Genetic Test Statistics
Pi
208.181871
Theta
177.087286
Tajima's D
0.643949
CLR
0.000249
CSRT
0.552072396380181
Interpretation
Uncertain
