Gene
KWMTBOMO15787
Pre Gene Modal
BGIBMGA000030
Annotation
PREDICTED:_alpha-sarcoglycan_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.462 Nuclear Reliability : 1.161
Sequence
CDS
ATGGCAAACGCACTGCTATGTTCGGCTTTGCTAACAGCGTGCGTGGTCTCCGCTCAGGTGCATACGGCCGTCGAAACAGAAATGTTCGCTATACCAATCAGCCCTAATTTATTTAATTGGACTTACCAAGAGTTCGACCAGCAGTACCGGTTTCACGCGTCGCTGCTGGGTAAGCCTGAGCTACCACAGTGGCTTCACTATATCTATAGTGGGCGACATCACTCTGGCTTCATATTCGGTACCCCACCCAGAGGCACCGAAGGACCTTTAACGCTGGAAGTAATCGGTTTGAATCGTCAAGATTTTGAGACGCGCCGCGTACTTTTGACACTGAAGGTCGAGAAGAAGGAGAAGATGGCCAGCCACGAAGTCGAGCTGAAGATCGACAACTTGAACGTCGAGGACTTGTTAGACGCACACCGGATGAATCGTTTGAAGGATATTCTGAGGACCAAACTCTGGACGGAAAGCAGTCAAGATCTGTACGCCACGTTCTTAGCTTCGGCCATAGATCTTGGTGCCAGATTGCCGCTGAAACCCAGCGACGGTGAGGGGCTGGTGGTCCGGCTGGGCAGTTCGCACCCGTTCTCGTCCGAACTGAAACGTCTCCGTGAGGAGGTGCGCCCCCTGTCGAAGCTGCCGAGCTGCCCTCGCGACTACAAGAGGACCACAGTCGAAAGACTCTTCAGGGACGCCGGCCTCACCCTTGACTGGTGCAGGTTTGAGCTGTTGAAGACGTACTACAAGCCCCGCTCGACCGTCCAGCTGGAGTACATGAGCGACACCCCGGCGGGGGGGCGGCCCGGGCGCGCCTCCCCCGCCCCCGCCCCCTGGTCGGCGCCGGCGCGGCACTCGCTGCCGACGCGGGGCTACGCGCGGCAACTATCCGCCGTCCTGGCCGCGCCCCTGGCTCTGCTGGCCGTGTCGGTCGCCGCCCTGACCGCGCTGGTCTGTTTCCATTACGCTGCTGTGAACGATCCAGAATCCGAGAGCTTCTTCGAAAACATATTTCACATTTGCATTGAATACAAACAAAGGAGGTTCGATAATTTGGACAAACGCTTTAATGTAATCTAA
Protein
MANALLCSALLTACVVSAQVHTAVETEMFAIPISPNLFNWTYQEFDQQYRFHASLLGKPELPQWLHYIYSGRHHSGFIFGTPPRGTEGPLTLEVIGLNRQDFETRRVLLTLKVEKKEKMASHEVELKIDNLNVEDLLDAHRMNRLKDILRTKLWTESSQDLYATFLASAIDLGARLPLKPSDGEGLVVRLGSSHPFSSELKRLREEVRPLSKLPSCPRDYKRTTVERLFRDAGLTLDWCRFELLKTYYKPRSTVQLEYMSDTPAGGRPGRASPAPAPWSAPARHSLPTRGYARQLSAVLAAPLALLAVSVAALTALVCFHYAAVNDPESESFFENIFHICIEYKQRRFDNLDKRFNVI
Summary
Uniprot
H9IS07
A0A0N1I6E3
A0A212FGL7
A0A0N0P9C1
A0A3S2NP66
A0A2W1BF29
+ More
A0A1W4WL98 A0A1Y1LMH7 A0A0T6AZN0 D6X1T3 A0A3L8DZW7 F4X4B4 A0A195BQR8 A0A151WR07 A0A158NXP4 A0A151IGS6 A0A195FGI7 A0A195E3Z2 A0A0A9X3D7 A0A0L7QKT0 E2BH32 E0VUX6 B3MPG8 A0A1W4V7Z9 A0A1I8M566 A0A1I8NTR1 A0A1A9WM28 W8CAK4 B4JBV8 A0A0L0CDJ9 Q9GT66 A0A0K8WFJ2 B4MWV3 A0A034WSQ7 B3N734 B4NWV5 A0A0J9QYK6 A0A1B0AEN3 Q9VLQ0 B4HYH9 A0A1B0GCJ3 M9PF78 B4KJM5 A0A0M4E9K5 B4LSH3 A0A0K8V1P1 A0A1A9VMG0 B4G905 T1PIE2 B4Q6P7 Q29MM9 A0A3B0K2H7 A0A1B0BL98 A0A182JW85 A0A182U276 A0A2S2QI50 A0A1A9Y3J8 A0A1S4FE33 A0A182NHD9 A0A1L8D8V4 A0A1L8D8X0 Q7PTB8 U5EQX6 A0A182IPH7
A0A1W4WL98 A0A1Y1LMH7 A0A0T6AZN0 D6X1T3 A0A3L8DZW7 F4X4B4 A0A195BQR8 A0A151WR07 A0A158NXP4 A0A151IGS6 A0A195FGI7 A0A195E3Z2 A0A0A9X3D7 A0A0L7QKT0 E2BH32 E0VUX6 B3MPG8 A0A1W4V7Z9 A0A1I8M566 A0A1I8NTR1 A0A1A9WM28 W8CAK4 B4JBV8 A0A0L0CDJ9 Q9GT66 A0A0K8WFJ2 B4MWV3 A0A034WSQ7 B3N734 B4NWV5 A0A0J9QYK6 A0A1B0AEN3 Q9VLQ0 B4HYH9 A0A1B0GCJ3 M9PF78 B4KJM5 A0A0M4E9K5 B4LSH3 A0A0K8V1P1 A0A1A9VMG0 B4G905 T1PIE2 B4Q6P7 Q29MM9 A0A3B0K2H7 A0A1B0BL98 A0A182JW85 A0A182U276 A0A2S2QI50 A0A1A9Y3J8 A0A1S4FE33 A0A182NHD9 A0A1L8D8V4 A0A1L8D8X0 Q7PTB8 U5EQX6 A0A182IPH7
Pubmed
19121390
26354079
22118469
28756777
28004739
18362917
+ More
19820115 30249741 21719571 21347285 25401762 20798317 20566863 17994087 25315136 24495485 26108605 11018515 25348373 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17510324 12364791 14747013 17210077
19820115 30249741 21719571 21347285 25401762 20798317 20566863 17994087 25315136 24495485 26108605 11018515 25348373 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17510324 12364791 14747013 17210077
EMBL
BABH01035559
KQ461188
KPJ07317.1
AGBW02008644
OWR52882.1
KQ459550
+ More
KPJ00009.1 RSAL01000019 RVE52751.1 KZ150146 PZC72901.1 GEZM01057630 JAV72207.1 LJIG01022531 KRT80087.1 KQ971371 EFA09965.1 QOIP01000002 RLU26001.1 GL888633 EGI58710.1 KQ976423 KYM88916.1 KQ982815 KYQ50237.1 ADTU01000477 ADTU01000478 ADTU01000479 ADTU01000480 KQ977671 KYN00639.1 KQ981606 KYN39508.1 KQ979685 KYN19801.1 GBHO01029095 GBRD01002079 JAG14509.1 JAG63742.1 KQ414940 KOC59222.1 GL448268 EFN84953.1 DS235797 EEB17182.1 CH902620 EDV31264.2 GAMC01005661 JAC00895.1 CH916368 EDW04061.1 JRES01000536 KNC30335.1 AF277391 AAG17400.1 GDHF01002378 JAI49936.1 CH963857 EDW76592.2 GAKP01001578 GAKP01001575 JAC57377.1 CH954177 EDV59330.2 CM000157 EDW88485.2 CM002910 KMY89031.1 AE014134 AY071690 AAF52634.1 AAL49312.1 CH480818 EDW52109.1 CCAG010021443 AGB92782.1 CH933807 EDW11470.2 CP012523 ALC38901.1 CH940649 EDW64795.2 GDHF01019789 JAI32525.1 CH479180 EDW28835.1 KA648444 AFP63073.1 CM000361 EDX04206.1 CH379060 EAL33664.2 OUUW01000004 SPP79806.1 JXJN01016271 GGMS01008188 MBY77391.1 GFDF01011185 JAV02899.1 GFDF01011186 JAV02898.1 AAAB01008807 EAA04148.5 GANO01004013 JAB55858.1
KPJ00009.1 RSAL01000019 RVE52751.1 KZ150146 PZC72901.1 GEZM01057630 JAV72207.1 LJIG01022531 KRT80087.1 KQ971371 EFA09965.1 QOIP01000002 RLU26001.1 GL888633 EGI58710.1 KQ976423 KYM88916.1 KQ982815 KYQ50237.1 ADTU01000477 ADTU01000478 ADTU01000479 ADTU01000480 KQ977671 KYN00639.1 KQ981606 KYN39508.1 KQ979685 KYN19801.1 GBHO01029095 GBRD01002079 JAG14509.1 JAG63742.1 KQ414940 KOC59222.1 GL448268 EFN84953.1 DS235797 EEB17182.1 CH902620 EDV31264.2 GAMC01005661 JAC00895.1 CH916368 EDW04061.1 JRES01000536 KNC30335.1 AF277391 AAG17400.1 GDHF01002378 JAI49936.1 CH963857 EDW76592.2 GAKP01001578 GAKP01001575 JAC57377.1 CH954177 EDV59330.2 CM000157 EDW88485.2 CM002910 KMY89031.1 AE014134 AY071690 AAF52634.1 AAL49312.1 CH480818 EDW52109.1 CCAG010021443 AGB92782.1 CH933807 EDW11470.2 CP012523 ALC38901.1 CH940649 EDW64795.2 GDHF01019789 JAI32525.1 CH479180 EDW28835.1 KA648444 AFP63073.1 CM000361 EDX04206.1 CH379060 EAL33664.2 OUUW01000004 SPP79806.1 JXJN01016271 GGMS01008188 MBY77391.1 GFDF01011185 JAV02899.1 GFDF01011186 JAV02898.1 AAAB01008807 EAA04148.5 GANO01004013 JAB55858.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000283053
UP000192223
+ More
UP000007266 UP000279307 UP000007755 UP000078540 UP000075809 UP000005205 UP000078542 UP000078541 UP000078492 UP000053825 UP000008237 UP000009046 UP000007801 UP000192221 UP000095301 UP000095300 UP000091820 UP000001070 UP000037069 UP000007798 UP000008711 UP000002282 UP000092445 UP000000803 UP000001292 UP000092444 UP000009192 UP000092553 UP000008792 UP000078200 UP000008744 UP000000304 UP000001819 UP000268350 UP000092460 UP000075881 UP000075902 UP000092443 UP000075884 UP000007062 UP000075880
UP000007266 UP000279307 UP000007755 UP000078540 UP000075809 UP000005205 UP000078542 UP000078541 UP000078492 UP000053825 UP000008237 UP000009046 UP000007801 UP000192221 UP000095301 UP000095300 UP000091820 UP000001070 UP000037069 UP000007798 UP000008711 UP000002282 UP000092445 UP000000803 UP000001292 UP000092444 UP000009192 UP000092553 UP000008792 UP000078200 UP000008744 UP000000304 UP000001819 UP000268350 UP000092460 UP000075881 UP000075902 UP000092443 UP000075884 UP000007062 UP000075880
Interpro
SUPFAM
SSF49313
SSF49313
Gene 3D
ProteinModelPortal
H9IS07
A0A0N1I6E3
A0A212FGL7
A0A0N0P9C1
A0A3S2NP66
A0A2W1BF29
+ More
A0A1W4WL98 A0A1Y1LMH7 A0A0T6AZN0 D6X1T3 A0A3L8DZW7 F4X4B4 A0A195BQR8 A0A151WR07 A0A158NXP4 A0A151IGS6 A0A195FGI7 A0A195E3Z2 A0A0A9X3D7 A0A0L7QKT0 E2BH32 E0VUX6 B3MPG8 A0A1W4V7Z9 A0A1I8M566 A0A1I8NTR1 A0A1A9WM28 W8CAK4 B4JBV8 A0A0L0CDJ9 Q9GT66 A0A0K8WFJ2 B4MWV3 A0A034WSQ7 B3N734 B4NWV5 A0A0J9QYK6 A0A1B0AEN3 Q9VLQ0 B4HYH9 A0A1B0GCJ3 M9PF78 B4KJM5 A0A0M4E9K5 B4LSH3 A0A0K8V1P1 A0A1A9VMG0 B4G905 T1PIE2 B4Q6P7 Q29MM9 A0A3B0K2H7 A0A1B0BL98 A0A182JW85 A0A182U276 A0A2S2QI50 A0A1A9Y3J8 A0A1S4FE33 A0A182NHD9 A0A1L8D8V4 A0A1L8D8X0 Q7PTB8 U5EQX6 A0A182IPH7
A0A1W4WL98 A0A1Y1LMH7 A0A0T6AZN0 D6X1T3 A0A3L8DZW7 F4X4B4 A0A195BQR8 A0A151WR07 A0A158NXP4 A0A151IGS6 A0A195FGI7 A0A195E3Z2 A0A0A9X3D7 A0A0L7QKT0 E2BH32 E0VUX6 B3MPG8 A0A1W4V7Z9 A0A1I8M566 A0A1I8NTR1 A0A1A9WM28 W8CAK4 B4JBV8 A0A0L0CDJ9 Q9GT66 A0A0K8WFJ2 B4MWV3 A0A034WSQ7 B3N734 B4NWV5 A0A0J9QYK6 A0A1B0AEN3 Q9VLQ0 B4HYH9 A0A1B0GCJ3 M9PF78 B4KJM5 A0A0M4E9K5 B4LSH3 A0A0K8V1P1 A0A1A9VMG0 B4G905 T1PIE2 B4Q6P7 Q29MM9 A0A3B0K2H7 A0A1B0BL98 A0A182JW85 A0A182U276 A0A2S2QI50 A0A1A9Y3J8 A0A1S4FE33 A0A182NHD9 A0A1L8D8V4 A0A1L8D8X0 Q7PTB8 U5EQX6 A0A182IPH7
Ontologies
GO
PANTHER
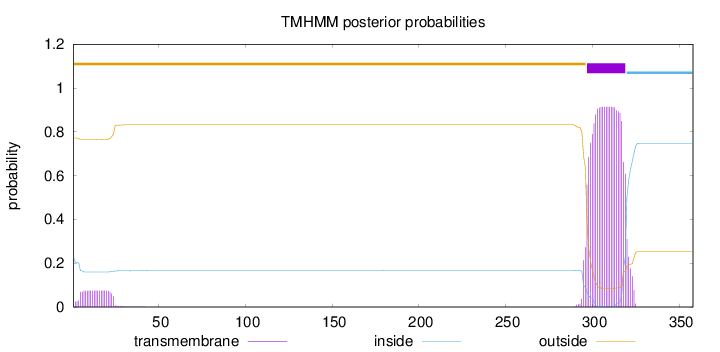
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.974021
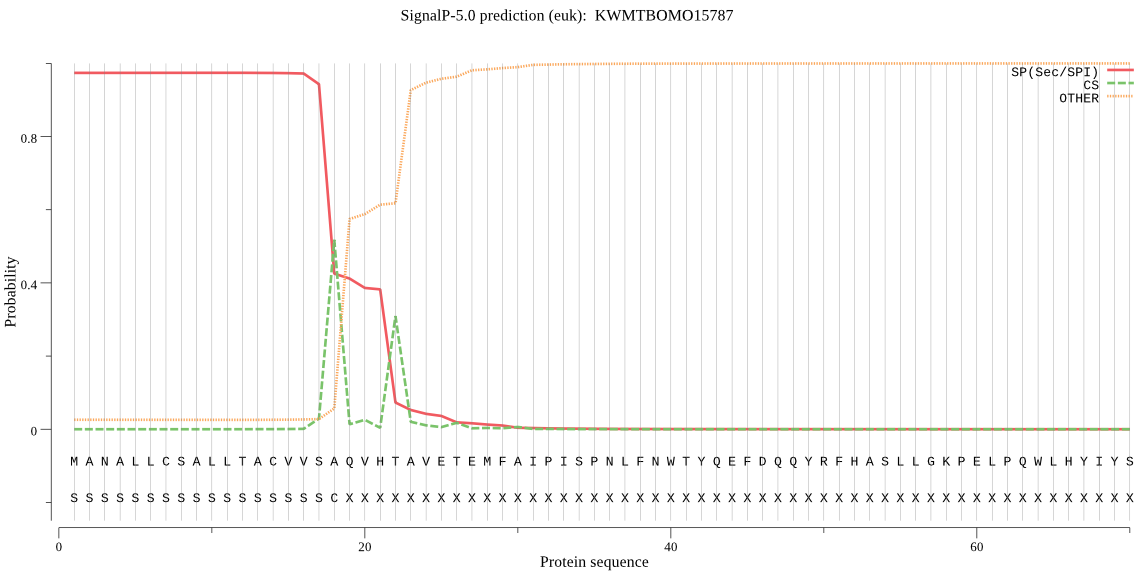
Length:
358
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.41321
Exp number, first 60 AAs:
1.56562
Total prob of N-in:
0.22846
outside
1 - 296
TMhelix
297 - 319
inside
320 - 358
Population Genetic Test Statistics
Pi
249.503472
Theta
21.100998
Tajima's D
-1.401023
CLR
114.274486
CSRT
0.0750462476876156
Interpretation
Uncertain
