Gene
KWMTBOMO15782
Annotation
PREDICTED:_cilia-_and_flagella-associated_protein_58-like_[Bombyx_mori]
Full name
Cilia- and flagella-associated protein 58
Alternative Name
Flagellum-associated protein 58
Location in the cell
Mitochondrial Reliability : 1.54 Nuclear Reliability : 1.46
Sequence
CDS
ATGTTGGTACTGAAAGATCGCGAACTGAAAGAGACCAGGAATTTATATTCTGCCGTCAAGGACATGCTGGCACTGCAGCCTAGTCCGGAAATACAGCAAACATTAAACAGAACACAGAGGGCGCTGGCTCAGCGAACATCTAAAATGAAGTGTCTCATTGCTGAACTAAGCATGCGCGAGAAACAAGCGACAGACCTTCGTCTGGAATTAGACCGTTTGAACGCGGATCTTCAATCGTACAAACAGAAATACTACGAGTTGAAGCGGGCACTCGACGCAGACGAAGCGAGAAGATTAAAAGTTCCGACGCCCGTCACAACTCCTGCTGCGCCGATTTCATAA
Protein
MLVLKDRELKETRNLYSAVKDMLALQPSPEIQQTLNRTQRALAQRTSKMKCLIAELSMREKQATDLRLELDRLNADLQSYKQKYYELKRALDADEARRLKVPTPVTTPAAPIS
Summary
Similarity
Belongs to the CFAP58 family.
Keywords
Cell projection
Cilium
Coiled coil
Flagellum
Feature
chain Cilia- and flagella-associated protein 58
Uniprot
A0A194QCE9
A0A0N1IBF0
A0A2A4J336
A0A212FPK9
A0A2W1BIL9
A0A3S2TQZ6
+ More
A0A1E1WLK4 A0A2H1W7H9 A0A310SQ36 A0A1Y1N8X6 A0A0C9QU87 A0A0G4IVH0 A0A3L8E0Q9 A0A026WEF8 A0A1W4WG87 E2ARW1 K7J7G6 J9J502 A0A078AEJ5 A0A0K6S8X7 A0A0J7KXP3 A0A2R5H2Q8 A0A139WBS4 A0A232EPU5 A0AUS3 A0A195C1N2 D7FKG1 A0A183P2R9 A8WFX1 A0A388KMS5 C1EH08 A0A1Y1IJ76 A0A2P8YEB4 F1QGB9 B8JHX6 C1MNZ6 A0A0K6S8L3 A0A061SN17 A0A1Y3NSL4 A0A1Q9EH45 A0A0M0JN86 A0A183R0A2 A0A1S3I8U9 A0A183MFT9 A0A3Q0KQX3 G4VF73 A0A183JZE3 A0A094ZPA8 A0A2G8L5A5 A0A2J7ZYL7 A0A1Y1VHC6 A8HUA7 A8HUA1 A0A150GBW4 V6LRH8 A0A0G4ELH9 A0A3M6TSA0 A0A2P6TGH8 A0A2H1C101 A0A0S4JDB4 G0V1Z5 A0A369S6H6 A0A0V0QW54 A0A2P6V590 A0A383VEV0 A0A060VRI1 A0A3P9ALP3 M3XI17 H3A1A5 G7YFR0 W5N277 A0EAT7 S9XSE0 A0A0D2K8T6 A0A176VCI6 A0A1A6GT85 A0A250WW83 A0A1Q9EU08 A0D6S5 A0A1S3L977 Q08D13 B2GU48 A0A397ER26 A0A3B4DM66 A0A024ULA7 A0A3R7WKI4 E1EWF3 A0A132NPU0 A0A397A227 A0A3R6YF08
A0A1E1WLK4 A0A2H1W7H9 A0A310SQ36 A0A1Y1N8X6 A0A0C9QU87 A0A0G4IVH0 A0A3L8E0Q9 A0A026WEF8 A0A1W4WG87 E2ARW1 K7J7G6 J9J502 A0A078AEJ5 A0A0K6S8X7 A0A0J7KXP3 A0A2R5H2Q8 A0A139WBS4 A0A232EPU5 A0AUS3 A0A195C1N2 D7FKG1 A0A183P2R9 A8WFX1 A0A388KMS5 C1EH08 A0A1Y1IJ76 A0A2P8YEB4 F1QGB9 B8JHX6 C1MNZ6 A0A0K6S8L3 A0A061SN17 A0A1Y3NSL4 A0A1Q9EH45 A0A0M0JN86 A0A183R0A2 A0A1S3I8U9 A0A183MFT9 A0A3Q0KQX3 G4VF73 A0A183JZE3 A0A094ZPA8 A0A2G8L5A5 A0A2J7ZYL7 A0A1Y1VHC6 A8HUA7 A8HUA1 A0A150GBW4 V6LRH8 A0A0G4ELH9 A0A3M6TSA0 A0A2P6TGH8 A0A2H1C101 A0A0S4JDB4 G0V1Z5 A0A369S6H6 A0A0V0QW54 A0A2P6V590 A0A383VEV0 A0A060VRI1 A0A3P9ALP3 M3XI17 H3A1A5 G7YFR0 W5N277 A0EAT7 S9XSE0 A0A0D2K8T6 A0A176VCI6 A0A1A6GT85 A0A250WW83 A0A1Q9EU08 A0D6S5 A0A1S3L977 Q08D13 B2GU48 A0A397ER26 A0A3B4DM66 A0A024ULA7 A0A3R7WKI4 E1EWF3 A0A132NPU0 A0A397A227 A0A3R6YF08
Pubmed
26354079
22118469
28756777
28004739
30249741
24508170
+ More
20798317 20075255 23382650 18362917 19820115 28648823 20520714 30007417 19359590 24865297 29403074 23594743 22253936 22246508 29023486 29294063 17932292 15998802 27102219 30382153 29178410 22331916 30042472 26486372 24755649 25069045 9215903 22023798 17086204 23149746 24373495 20929575 26003141
20798317 20075255 23382650 18362917 19820115 28648823 20520714 30007417 19359590 24865297 29403074 23594743 22253936 22246508 29023486 29294063 17932292 15998802 27102219 30382153 29178410 22331916 30042472 26486372 24755649 25069045 9215903 22023798 17086204 23149746 24373495 20929575 26003141
EMBL
KQ459193
KPJ03142.1
KQ461188
KPJ07319.1
NWSH01003568
PCG66098.1
+ More
AGBW02001704 OWR55688.1 KZ150129 PZC73077.1 RSAL01000019 RVE52753.1 GDQN01003199 JAT87855.1 ODYU01006824 SOQ49030.1 KQ760240 OAD61303.1 GEZM01014690 JAV92037.1 GBYB01004227 JAG73994.1 CDSF01000090 OVEO01000007 CEO99227.1 SPQ97237.1 QOIP01000001 RLU26304.1 KK107260 EZA54046.1 GL442192 EFN63810.1 AAZX01003544 AMCR01003991 EJY84478.1 CCKQ01008801 CDW80266.1 CDMZ01002235 CUC10000.1 LBMM01002258 KMQ95066.1 BEYU01000209 GBG34684.1 KQ971372 KYB25376.1 NNAY01002899 OXU20358.1 BC124828 AAI24829.1 KQ978379 KYM94520.1 FN648026 CBJ29363.1 UZAL01029083 VDP45825.1 BC154489 AAI54490.1 BFEA01000145 GBG71364.1 CP001332 ACO67215.1 DF237406 GAQ88766.1 PYGN01000665 PSN42585.1 CR381623 CU407322 LO018637 GG663737 EEH58843.1 CUC10001.1 GBEZ01025124 GBEZ01023309 GBEZ01000620 JAC61928.1 JAC63570.1 JAC84280.1 KZ150777 OUM68143.1 LSRX01000154 OLQ06698.1 JWZX01002665 KOO27787.1 UZAI01016845 VDP17023.1 HE601626 CCD78581.1 UZAK01032601 VDP29247.1 KL250637 KGB34839.1 MRZV01000214 PIK55459.1 PGGS01000312 PNH05361.1 MCFH01000008 ORX55851.1 DS496109 CM008974 EDP09093.1 PNW74075.1 EDP08857.1 LSYV01000037 KXZ47347.1 KI546046 EST47165.1 CDMY01000256 CEL97807.1 RCHS01003041 RMX44265.1 LHPG02000017 PRW33224.1 KZ431889 PIS81114.1 CYKH01001733 CUG89385.1 HE575324 CCC95667.1 NOWV01000056 RDD42359.1 LDAU01000096 KRX06422.1 LHPF02000028 PSC69261.1 FNXT01000270 SZX62906.1 FR904289 CDQ57558.1 AFYH01068381 AFYH01068382 AFYH01068383 AFYH01068384 AFYH01068385 AFYH01068386 AFYH01068387 AFYH01068388 AFYH01068389 AFYH01068390 DF143201 GAA51793.1 AHAT01017371 CT868668 CAK92404.1 KB018013 EPY74615.1 KK100353 KIZ06593.1 LVLJ01004028 OAE18619.1 LZPO01068676 OBS69543.1 BEGY01000010 GAX75026.1 LSRX01000069 OLQ10911.1 CT868318 CAK78742.1 BC123995 AAI23996.1 BC166135 AAI66135.1 QUTE01017752 RHY91835.1 KI913955 ETW07089.1 MZMZ02002624 RQM25021.1 ACVC01000028 EFO65452.1 JXTI01000140 KWX12028.1 QUTA01010510 QUSZ01007742 QUTC01004564 QUTD01003468 QUTH01003381 QUTF01009957 RHX99539.1 RHY01672.1 RHY63797.1 RHY72731.1 RHZ19761.1 RHZ33735.1 QUTB01012455 RHY33878.1
AGBW02001704 OWR55688.1 KZ150129 PZC73077.1 RSAL01000019 RVE52753.1 GDQN01003199 JAT87855.1 ODYU01006824 SOQ49030.1 KQ760240 OAD61303.1 GEZM01014690 JAV92037.1 GBYB01004227 JAG73994.1 CDSF01000090 OVEO01000007 CEO99227.1 SPQ97237.1 QOIP01000001 RLU26304.1 KK107260 EZA54046.1 GL442192 EFN63810.1 AAZX01003544 AMCR01003991 EJY84478.1 CCKQ01008801 CDW80266.1 CDMZ01002235 CUC10000.1 LBMM01002258 KMQ95066.1 BEYU01000209 GBG34684.1 KQ971372 KYB25376.1 NNAY01002899 OXU20358.1 BC124828 AAI24829.1 KQ978379 KYM94520.1 FN648026 CBJ29363.1 UZAL01029083 VDP45825.1 BC154489 AAI54490.1 BFEA01000145 GBG71364.1 CP001332 ACO67215.1 DF237406 GAQ88766.1 PYGN01000665 PSN42585.1 CR381623 CU407322 LO018637 GG663737 EEH58843.1 CUC10001.1 GBEZ01025124 GBEZ01023309 GBEZ01000620 JAC61928.1 JAC63570.1 JAC84280.1 KZ150777 OUM68143.1 LSRX01000154 OLQ06698.1 JWZX01002665 KOO27787.1 UZAI01016845 VDP17023.1 HE601626 CCD78581.1 UZAK01032601 VDP29247.1 KL250637 KGB34839.1 MRZV01000214 PIK55459.1 PGGS01000312 PNH05361.1 MCFH01000008 ORX55851.1 DS496109 CM008974 EDP09093.1 PNW74075.1 EDP08857.1 LSYV01000037 KXZ47347.1 KI546046 EST47165.1 CDMY01000256 CEL97807.1 RCHS01003041 RMX44265.1 LHPG02000017 PRW33224.1 KZ431889 PIS81114.1 CYKH01001733 CUG89385.1 HE575324 CCC95667.1 NOWV01000056 RDD42359.1 LDAU01000096 KRX06422.1 LHPF02000028 PSC69261.1 FNXT01000270 SZX62906.1 FR904289 CDQ57558.1 AFYH01068381 AFYH01068382 AFYH01068383 AFYH01068384 AFYH01068385 AFYH01068386 AFYH01068387 AFYH01068388 AFYH01068389 AFYH01068390 DF143201 GAA51793.1 AHAT01017371 CT868668 CAK92404.1 KB018013 EPY74615.1 KK100353 KIZ06593.1 LVLJ01004028 OAE18619.1 LZPO01068676 OBS69543.1 BEGY01000010 GAX75026.1 LSRX01000069 OLQ10911.1 CT868318 CAK78742.1 BC123995 AAI23996.1 BC166135 AAI66135.1 QUTE01017752 RHY91835.1 KI913955 ETW07089.1 MZMZ02002624 RQM25021.1 ACVC01000028 EFO65452.1 JXTI01000140 KWX12028.1 QUTA01010510 QUSZ01007742 QUTC01004564 QUTD01003468 QUTH01003381 QUTF01009957 RHX99539.1 RHY01672.1 RHY63797.1 RHY72731.1 RHZ19761.1 RHZ33735.1 QUTB01012455 RHY33878.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000007151
UP000283053
UP000039324
+ More
UP000290189 UP000279307 UP000053097 UP000192223 UP000000311 UP000002358 UP000036403 UP000007266 UP000215335 UP000078542 UP000002630 UP000050791 UP000265515 UP000002009 UP000054558 UP000245037 UP000000437 UP000001876 UP000195826 UP000050792 UP000085678 UP000050790 UP000277204 UP000008854 UP000050789 UP000230750 UP000193719 UP000006906 UP000075714 UP000041254 UP000275408 UP000239899 UP000253843 UP000239649 UP000256970 UP000193380 UP000265140 UP000008672 UP000018468 UP000000600 UP000054498 UP000092124 UP000232323 UP000087266 UP000261440 UP000024375 UP000008974
UP000290189 UP000279307 UP000053097 UP000192223 UP000000311 UP000002358 UP000036403 UP000007266 UP000215335 UP000078542 UP000002630 UP000050791 UP000265515 UP000002009 UP000054558 UP000245037 UP000000437 UP000001876 UP000195826 UP000050792 UP000085678 UP000050790 UP000277204 UP000008854 UP000050789 UP000230750 UP000193719 UP000006906 UP000075714 UP000041254 UP000275408 UP000239899 UP000253843 UP000239649 UP000256970 UP000193380 UP000265140 UP000008672 UP000018468 UP000000600 UP000054498 UP000092124 UP000232323 UP000087266 UP000261440 UP000024375 UP000008974
Interpro
Gene 3D
ProteinModelPortal
A0A194QCE9
A0A0N1IBF0
A0A2A4J336
A0A212FPK9
A0A2W1BIL9
A0A3S2TQZ6
+ More
A0A1E1WLK4 A0A2H1W7H9 A0A310SQ36 A0A1Y1N8X6 A0A0C9QU87 A0A0G4IVH0 A0A3L8E0Q9 A0A026WEF8 A0A1W4WG87 E2ARW1 K7J7G6 J9J502 A0A078AEJ5 A0A0K6S8X7 A0A0J7KXP3 A0A2R5H2Q8 A0A139WBS4 A0A232EPU5 A0AUS3 A0A195C1N2 D7FKG1 A0A183P2R9 A8WFX1 A0A388KMS5 C1EH08 A0A1Y1IJ76 A0A2P8YEB4 F1QGB9 B8JHX6 C1MNZ6 A0A0K6S8L3 A0A061SN17 A0A1Y3NSL4 A0A1Q9EH45 A0A0M0JN86 A0A183R0A2 A0A1S3I8U9 A0A183MFT9 A0A3Q0KQX3 G4VF73 A0A183JZE3 A0A094ZPA8 A0A2G8L5A5 A0A2J7ZYL7 A0A1Y1VHC6 A8HUA7 A8HUA1 A0A150GBW4 V6LRH8 A0A0G4ELH9 A0A3M6TSA0 A0A2P6TGH8 A0A2H1C101 A0A0S4JDB4 G0V1Z5 A0A369S6H6 A0A0V0QW54 A0A2P6V590 A0A383VEV0 A0A060VRI1 A0A3P9ALP3 M3XI17 H3A1A5 G7YFR0 W5N277 A0EAT7 S9XSE0 A0A0D2K8T6 A0A176VCI6 A0A1A6GT85 A0A250WW83 A0A1Q9EU08 A0D6S5 A0A1S3L977 Q08D13 B2GU48 A0A397ER26 A0A3B4DM66 A0A024ULA7 A0A3R7WKI4 E1EWF3 A0A132NPU0 A0A397A227 A0A3R6YF08
A0A1E1WLK4 A0A2H1W7H9 A0A310SQ36 A0A1Y1N8X6 A0A0C9QU87 A0A0G4IVH0 A0A3L8E0Q9 A0A026WEF8 A0A1W4WG87 E2ARW1 K7J7G6 J9J502 A0A078AEJ5 A0A0K6S8X7 A0A0J7KXP3 A0A2R5H2Q8 A0A139WBS4 A0A232EPU5 A0AUS3 A0A195C1N2 D7FKG1 A0A183P2R9 A8WFX1 A0A388KMS5 C1EH08 A0A1Y1IJ76 A0A2P8YEB4 F1QGB9 B8JHX6 C1MNZ6 A0A0K6S8L3 A0A061SN17 A0A1Y3NSL4 A0A1Q9EH45 A0A0M0JN86 A0A183R0A2 A0A1S3I8U9 A0A183MFT9 A0A3Q0KQX3 G4VF73 A0A183JZE3 A0A094ZPA8 A0A2G8L5A5 A0A2J7ZYL7 A0A1Y1VHC6 A8HUA7 A8HUA1 A0A150GBW4 V6LRH8 A0A0G4ELH9 A0A3M6TSA0 A0A2P6TGH8 A0A2H1C101 A0A0S4JDB4 G0V1Z5 A0A369S6H6 A0A0V0QW54 A0A2P6V590 A0A383VEV0 A0A060VRI1 A0A3P9ALP3 M3XI17 H3A1A5 G7YFR0 W5N277 A0EAT7 S9XSE0 A0A0D2K8T6 A0A176VCI6 A0A1A6GT85 A0A250WW83 A0A1Q9EU08 A0D6S5 A0A1S3L977 Q08D13 B2GU48 A0A397ER26 A0A3B4DM66 A0A024ULA7 A0A3R7WKI4 E1EWF3 A0A132NPU0 A0A397A227 A0A3R6YF08
Ontologies
GO
PANTHER
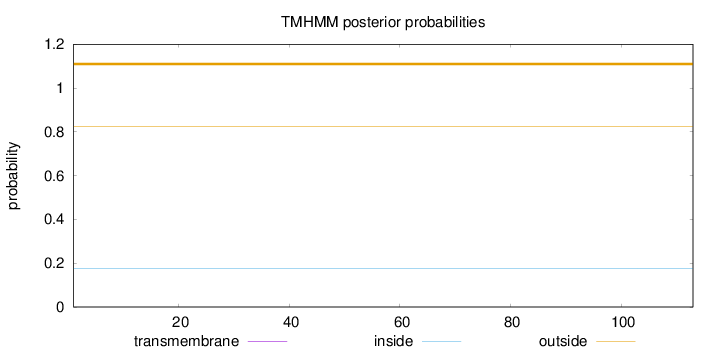
Topology
Subcellular location
Length:
113
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.17540
outside
1 - 113
Population Genetic Test Statistics
Pi
15.252811
Theta
15.194702
Tajima's D
0.436099
CLR
0.760621
CSRT
0.508324583770811
Interpretation
Uncertain
