Gene
KWMTBOMO15781 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002346
Annotation
PREDICTED:_C-terminal-binding_protein_isoform_X1_[Bombyx_mori]
Full name
C-terminal-binding protein
Alternative Name
dCtBP
Location in the cell
Cytoplasmic Reliability : 1.091 Mitochondrial Reliability : 1.556
Sequence
CDS
ATGGACAAACGCAAGATGCTGCCAAAGAGAGCGCGCATGGATAGTATGCGGGGTCCTATTGCAAACGGACCCCTGCAATCAAGGCCACTCGTGGCCCTGCTGGACGGTCGGGATTGTACCGTCGAGATGCCGATCCTGAAAGACGTGGCGACGGTCGCCTTCTGCGACGCGCAGTCGACGTCCGAAATACACGAGAAGGTACTCAACGAAGCGGTCGGGGCCCTCATGTGGCACACCATTATACTAACCAAGGAGGACCTTGAGAAGTTCAAGGCGTTACGGATCATAGTACGCATCGGGTCAGGTGTCGATAATATCGACGTTAAAGCCGCCGGCGAATTAGGAATAGCGGTATGTAATGTTCCCGGGTACGGAGTCGAGGAGGTAGCCGACACCACGATGTGTCTCATATTGAATTTATATCGACGGACGTATTGGCTAGCCAATATGGTCAGAGAAGGAAAAAAATTTACAGGGCCGGAGCAGGTGCGCGAGGCGGCGGCGGGGTGCGCGCGCATCCGCGGCGACACGCTCGGCATCGTGGGGCTCGGCCGGATCGGCTCGGCCGTGGCGCTGCGGGCCAAGGCGTTCGGCTTCAATGTCATATTCTACGACCCCTACCTGCCTGACGGCATCGAGAAGTCGCTGGGCCTCACCAGGGTCTACACGCTACAGGACTTATTATTCCAGAGTGACTGTGTCTCACTGCACTGCAGCTTAAACGAGCACAATCACCACCTCATCAACGAGTTCACCATCAAACAGATGCGTCCAGGTGCGTTTCTGGTGAACACGGCGCGCGGCGGGCTCGTGGACGACGAGGGGCTCGCCGCCGCGCTCAAGCAGGGCCGCATCCGAGCCGCCGCGCTAGACGTGCACGAGAACGAACCCTTCAACGTCTTCCAGCAGGGTCCGCTGAAAGACGCTCCGAACCTGCTGTGCACGCCGCACGCGGCCTTCTACTCGGACGCGTCCGCACAGGAGCTGCGCGAGATGGCCGCCTCGGAGATCCGCCGCGCCATCGTCGGCCGCATCCCCGACTGTCTGCGGAACTGCGTCAATAAGGACTACTTCATGGCGGGAACGGGCGGCGTGGTGGGCGGAGTGGTGGGCGGCGTGGTGGGCGGCGTGGTGTCGGGGTCGGGCGGCGCGGGCGGCGCGTACGGCGAGGGCATCAACGGCGGGTACTACGGCGCCGCGCAGGCCGCGCACTCCACCACCGCCGTGCACGAGCCCGCCGCGCACGCCGCGCAGCCGCCGCCCGCGCAGCCCGCGCCGCAGCAGCAGCCGCCCATCACGCTGCCCATAAACACGACGGACCCGGCCAACCACCAGATCCCCAAGCAGGAGAGCTCGGACGTGCACTAG
Protein
MDKRKMLPKRARMDSMRGPIANGPLQSRPLVALLDGRDCTVEMPILKDVATVAFCDAQSTSEIHEKVLNEAVGALMWHTIILTKEDLEKFKALRIIVRIGSGVDNIDVKAAGELGIAVCNVPGYGVEEVADTTMCLILNLYRRTYWLANMVREGKKFTGPEQVREAAAGCARIRGDTLGIVGLGRIGSAVALRAKAFGFNVIFYDPYLPDGIEKSLGLTRVYTLQDLLFQSDCVSLHCSLNEHNHHLINEFTIKQMRPGAFLVNTARGGLVDDEGLAAALKQGRIRAAALDVHENEPFNVFQQGPLKDAPNLLCTPHAAFYSDASAQELREMAASEIRRAIVGRIPDCLRNCVNKDYFMAGTGGVVGGVVGGVVGGVVSGSGGAGGAYGEGINGGYYGAAQAAHSTTAVHEPAAHAAQPPPAQPAPQQQPPITLPINTTDPANHQIPKQESSDVH
Summary
Description
Corepressor targeting diverse transcription regulators. Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression.
Subunit
Homodimer. Interacts with hairy (h), knirps (kni), snail (sna), and Enhancer of split m-delta (HLHm-delta). Complex may be involved in transcriptional repression. Interacts also with adenovirus E1A protein.
Similarity
Belongs to the D-isomer specific 2-hydroxyacid dehydrogenase family.
Keywords
Alternative splicing
Complete proteome
Nucleus
Oxidoreductase
Reference proteome
Repressor
Transcription
Transcription regulation
Feature
chain C-terminal-binding protein
splice variant In isoform A.
splice variant In isoform A.
Uniprot
A0A3S2M7F4
A0A212ERH5
H9IYL2
A0A2H1W7H4
A0A222AJF1
S4P1X1
+ More
A0A2W1BDW3 A0A194QC77 A0A1B6F6H4 A0A1B6DLE2 K7INJ3 A0A1B6LTR7 E2ADM8 A0A0N0U6Q9 A0A2J7PI32 V9I9U6 A0A067RB42 A0A0A9YD33 A0A087ZWM2 V9IC45 A0A026WZT5 A0A3L8DRR7 A0A151WRX6 A0A1Y1M9L8 A0A158NPF3 A0A224XC60 A0A310SG02 A0A069DUX2 A0A023F5Q7 A0A0P4VTC3 A0A154PLQ9 A0A0A9XCJ8 A0A0V0G9V3 E0W0A8 U4UHK9 A0A1B6DCA0 N6U4A8 A0A0C9RBU0 A0A1W4WUN9 A0A1B6EC23 A0A1W4X5D5 A0A0A9Y317 A0A0A9Y8M3 A0A0A9Y5K9 A0A1S3CWX9 A0A0A9Y744 A0A0A9Z857 A0A0A9Z291 A0A1S3CVK8 A0A1S3CVR8 V5GVC8 D6X2E0 A0A2A3E232 A0A195E543 A0A1W4WUM5 A0A1W4X5J2 A0A1B0GH93 A0A1I8N265 A0A1I8QDQ6 A0A1I8QDN8 A0A1I8N267 A0A0L0C742 A0A1A9XRG3 A0A1A9UFQ6 A0A1B0BVU9 A0A1A9ZA08 A0A1A9WS45 A0A1B0FIQ7 A0A0T6B5T9 A0A2S2QVF1 A0A1L8E0U4 A0A1L8E0P4 A0A1I8N289 U5ETF2 A0A1I8QDQ7 A0A195BA66 J9JJP6 B4N9Y3 Q299Q6 B4G5K5 A0A2S2NDF5 A0A2H8U079 A0A0Q9WYB4 B4K8I6 B4PPV3 B4HGC4 B3P196 A0A0A1WIH5 A0A1W4U824 A0A1W4UJ87 B4R1X7 O46036 A0A0K8VGV1 A0A0R1E2T5 A0A0A1WV51 W8AQ12 A0A0R1E2U6 A0A0M4EVQ0 B4JEW9 A0A0K8U655
A0A2W1BDW3 A0A194QC77 A0A1B6F6H4 A0A1B6DLE2 K7INJ3 A0A1B6LTR7 E2ADM8 A0A0N0U6Q9 A0A2J7PI32 V9I9U6 A0A067RB42 A0A0A9YD33 A0A087ZWM2 V9IC45 A0A026WZT5 A0A3L8DRR7 A0A151WRX6 A0A1Y1M9L8 A0A158NPF3 A0A224XC60 A0A310SG02 A0A069DUX2 A0A023F5Q7 A0A0P4VTC3 A0A154PLQ9 A0A0A9XCJ8 A0A0V0G9V3 E0W0A8 U4UHK9 A0A1B6DCA0 N6U4A8 A0A0C9RBU0 A0A1W4WUN9 A0A1B6EC23 A0A1W4X5D5 A0A0A9Y317 A0A0A9Y8M3 A0A0A9Y5K9 A0A1S3CWX9 A0A0A9Y744 A0A0A9Z857 A0A0A9Z291 A0A1S3CVK8 A0A1S3CVR8 V5GVC8 D6X2E0 A0A2A3E232 A0A195E543 A0A1W4WUM5 A0A1W4X5J2 A0A1B0GH93 A0A1I8N265 A0A1I8QDQ6 A0A1I8QDN8 A0A1I8N267 A0A0L0C742 A0A1A9XRG3 A0A1A9UFQ6 A0A1B0BVU9 A0A1A9ZA08 A0A1A9WS45 A0A1B0FIQ7 A0A0T6B5T9 A0A2S2QVF1 A0A1L8E0U4 A0A1L8E0P4 A0A1I8N289 U5ETF2 A0A1I8QDQ7 A0A195BA66 J9JJP6 B4N9Y3 Q299Q6 B4G5K5 A0A2S2NDF5 A0A2H8U079 A0A0Q9WYB4 B4K8I6 B4PPV3 B4HGC4 B3P196 A0A0A1WIH5 A0A1W4U824 A0A1W4UJ87 B4R1X7 O46036 A0A0K8VGV1 A0A0R1E2T5 A0A0A1WV51 W8AQ12 A0A0R1E2U6 A0A0M4EVQ0 B4JEW9 A0A0K8U655
Pubmed
22118469
19121390
23622113
28756777
26354079
20075255
+ More
20798317 24845553 25401762 24508170 30249741 28004739 21347285 26334808 25474469 27129103 20566863 23537049 26823975 18362917 19820115 25315136 26108605 17994087 15632085 17550304 25830018 9524128 9525852 10731132 12537572 12537569 24495485
20798317 24845553 25401762 24508170 30249741 28004739 21347285 26334808 25474469 27129103 20566863 23537049 26823975 18362917 19820115 25315136 26108605 17994087 15632085 17550304 25830018 9524128 9525852 10731132 12537572 12537569 24495485
EMBL
RSAL01000019
RVE52754.1
AGBW02013027
OWR44069.1
BABH01020914
BABH01020915
+ More
BABH01020916 ODYU01006824 SOQ49031.1 KY609804 ASO76495.1 GAIX01012280 JAA80280.1 KZ150129 PZC73078.1 KQ459193 KPJ03143.1 GECZ01024316 JAS45453.1 GEDC01010813 JAS26485.1 GEBQ01023355 GEBQ01012905 JAT16622.1 JAT27072.1 GL438820 EFN68390.1 KQ435721 KOX78528.1 NEVH01025129 PNF16000.1 JR037648 JR037649 AEY57892.1 AEY57893.1 KK852766 KDR16976.1 GBHO01016159 GBRD01012340 JAG27445.1 JAG53484.1 JR037650 JR037651 AEY57894.1 AEY57895.1 KK107063 EZA61256.1 QOIP01000005 RLU22863.1 KQ982807 KYQ50425.1 GEZM01036900 GEZM01036899 JAV82353.1 ADTU01022481 GFTR01006431 JAW09995.1 KQ768431 OAD53169.1 GBGD01001397 JAC87492.1 GBBI01002209 JAC16503.1 GDKW01000119 JAI56476.1 KQ434938 KZC12160.1 GBHO01028789 JAG14815.1 GECL01001439 JAP04685.1 DS235858 EEB19064.1 KB632204 ERL90101.1 GEDC01013975 JAS23323.1 APGK01043400 APGK01043401 APGK01043402 KB741014 ENN75461.1 GBYB01012614 GBYB01014044 GBYB01014052 JAG82381.1 JAG83811.1 JAG83819.1 GEDC01001810 JAS35488.1 GBHO01016152 JAG27452.1 GBHO01016156 GBRD01012339 GDHC01021047 JAG27448.1 JAG53485.1 JAP97581.1 GBHO01016155 GDHC01013700 JAG27449.1 JAQ04929.1 GBHO01016158 GBRD01012338 GDHC01000554 JAG27446.1 JAG53486.1 JAQ18075.1 GBHO01005609 GDHC01018275 JAG37995.1 JAQ00354.1 GBHO01005618 JAG37986.1 GALX01002834 JAB65632.1 KQ971371 EFA10253.2 KZ288438 PBC25748.1 KQ979608 KYN20305.1 AJWK01004097 AJWK01004098 AJWK01004099 AJWK01004100 AJWK01004101 JRES01000809 KNC28223.1 JXJN01021482 CCAG010023921 LJIG01009602 KRT82751.1 GGMS01011899 MBY81102.1 GFDF01001785 JAV12299.1 GFDF01001786 JAV12298.1 GANO01001948 JAB57923.1 KQ976542 KYM81120.1 ABLF02024982 CH964232 EDW81738.1 CM000070 EAL27645.2 CH479179 EDW24871.1 GGMR01002550 MBY15169.1 GFXV01008138 MBW19943.1 CH933806 KRG00991.1 EDW14385.1 CM000160 EDW97180.1 CH480815 EDW42372.1 CH954181 EDV49285.1 GBXI01015605 JAC98686.1 CM000364 EDX13129.1 AJ224690 AB011840 AE014297 AY060646 AY069170 GDHF01014569 JAI37745.1 KRK03563.1 GBXI01011776 GBXI01008114 JAD02516.1 JAD06178.1 GAMC01015686 GAMC01015685 GAMC01015684 GAMC01015683 JAB90870.1 KRK03565.1 CP012526 ALC47455.1 CH916369 EDV93250.1 GDHF01030278 GDHF01013710 JAI22036.1 JAI38604.1
BABH01020916 ODYU01006824 SOQ49031.1 KY609804 ASO76495.1 GAIX01012280 JAA80280.1 KZ150129 PZC73078.1 KQ459193 KPJ03143.1 GECZ01024316 JAS45453.1 GEDC01010813 JAS26485.1 GEBQ01023355 GEBQ01012905 JAT16622.1 JAT27072.1 GL438820 EFN68390.1 KQ435721 KOX78528.1 NEVH01025129 PNF16000.1 JR037648 JR037649 AEY57892.1 AEY57893.1 KK852766 KDR16976.1 GBHO01016159 GBRD01012340 JAG27445.1 JAG53484.1 JR037650 JR037651 AEY57894.1 AEY57895.1 KK107063 EZA61256.1 QOIP01000005 RLU22863.1 KQ982807 KYQ50425.1 GEZM01036900 GEZM01036899 JAV82353.1 ADTU01022481 GFTR01006431 JAW09995.1 KQ768431 OAD53169.1 GBGD01001397 JAC87492.1 GBBI01002209 JAC16503.1 GDKW01000119 JAI56476.1 KQ434938 KZC12160.1 GBHO01028789 JAG14815.1 GECL01001439 JAP04685.1 DS235858 EEB19064.1 KB632204 ERL90101.1 GEDC01013975 JAS23323.1 APGK01043400 APGK01043401 APGK01043402 KB741014 ENN75461.1 GBYB01012614 GBYB01014044 GBYB01014052 JAG82381.1 JAG83811.1 JAG83819.1 GEDC01001810 JAS35488.1 GBHO01016152 JAG27452.1 GBHO01016156 GBRD01012339 GDHC01021047 JAG27448.1 JAG53485.1 JAP97581.1 GBHO01016155 GDHC01013700 JAG27449.1 JAQ04929.1 GBHO01016158 GBRD01012338 GDHC01000554 JAG27446.1 JAG53486.1 JAQ18075.1 GBHO01005609 GDHC01018275 JAG37995.1 JAQ00354.1 GBHO01005618 JAG37986.1 GALX01002834 JAB65632.1 KQ971371 EFA10253.2 KZ288438 PBC25748.1 KQ979608 KYN20305.1 AJWK01004097 AJWK01004098 AJWK01004099 AJWK01004100 AJWK01004101 JRES01000809 KNC28223.1 JXJN01021482 CCAG010023921 LJIG01009602 KRT82751.1 GGMS01011899 MBY81102.1 GFDF01001785 JAV12299.1 GFDF01001786 JAV12298.1 GANO01001948 JAB57923.1 KQ976542 KYM81120.1 ABLF02024982 CH964232 EDW81738.1 CM000070 EAL27645.2 CH479179 EDW24871.1 GGMR01002550 MBY15169.1 GFXV01008138 MBW19943.1 CH933806 KRG00991.1 EDW14385.1 CM000160 EDW97180.1 CH480815 EDW42372.1 CH954181 EDV49285.1 GBXI01015605 JAC98686.1 CM000364 EDX13129.1 AJ224690 AB011840 AE014297 AY060646 AY069170 GDHF01014569 JAI37745.1 KRK03563.1 GBXI01011776 GBXI01008114 JAD02516.1 JAD06178.1 GAMC01015686 GAMC01015685 GAMC01015684 GAMC01015683 JAB90870.1 KRK03565.1 CP012526 ALC47455.1 CH916369 EDV93250.1 GDHF01030278 GDHF01013710 JAI22036.1 JAI38604.1
Proteomes
UP000283053
UP000007151
UP000005204
UP000053268
UP000002358
UP000000311
+ More
UP000053105 UP000235965 UP000027135 UP000005203 UP000053097 UP000279307 UP000075809 UP000005205 UP000076502 UP000009046 UP000030742 UP000019118 UP000192223 UP000079169 UP000007266 UP000242457 UP000078492 UP000092461 UP000095301 UP000095300 UP000037069 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000092444 UP000078540 UP000007819 UP000007798 UP000001819 UP000008744 UP000009192 UP000002282 UP000001292 UP000008711 UP000192221 UP000000304 UP000000803 UP000092553 UP000001070
UP000053105 UP000235965 UP000027135 UP000005203 UP000053097 UP000279307 UP000075809 UP000005205 UP000076502 UP000009046 UP000030742 UP000019118 UP000192223 UP000079169 UP000007266 UP000242457 UP000078492 UP000092461 UP000095301 UP000095300 UP000037069 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000092444 UP000078540 UP000007819 UP000007798 UP000001819 UP000008744 UP000009192 UP000002282 UP000001292 UP000008711 UP000192221 UP000000304 UP000000803 UP000092553 UP000001070
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
A0A3S2M7F4
A0A212ERH5
H9IYL2
A0A2H1W7H4
A0A222AJF1
S4P1X1
+ More
A0A2W1BDW3 A0A194QC77 A0A1B6F6H4 A0A1B6DLE2 K7INJ3 A0A1B6LTR7 E2ADM8 A0A0N0U6Q9 A0A2J7PI32 V9I9U6 A0A067RB42 A0A0A9YD33 A0A087ZWM2 V9IC45 A0A026WZT5 A0A3L8DRR7 A0A151WRX6 A0A1Y1M9L8 A0A158NPF3 A0A224XC60 A0A310SG02 A0A069DUX2 A0A023F5Q7 A0A0P4VTC3 A0A154PLQ9 A0A0A9XCJ8 A0A0V0G9V3 E0W0A8 U4UHK9 A0A1B6DCA0 N6U4A8 A0A0C9RBU0 A0A1W4WUN9 A0A1B6EC23 A0A1W4X5D5 A0A0A9Y317 A0A0A9Y8M3 A0A0A9Y5K9 A0A1S3CWX9 A0A0A9Y744 A0A0A9Z857 A0A0A9Z291 A0A1S3CVK8 A0A1S3CVR8 V5GVC8 D6X2E0 A0A2A3E232 A0A195E543 A0A1W4WUM5 A0A1W4X5J2 A0A1B0GH93 A0A1I8N265 A0A1I8QDQ6 A0A1I8QDN8 A0A1I8N267 A0A0L0C742 A0A1A9XRG3 A0A1A9UFQ6 A0A1B0BVU9 A0A1A9ZA08 A0A1A9WS45 A0A1B0FIQ7 A0A0T6B5T9 A0A2S2QVF1 A0A1L8E0U4 A0A1L8E0P4 A0A1I8N289 U5ETF2 A0A1I8QDQ7 A0A195BA66 J9JJP6 B4N9Y3 Q299Q6 B4G5K5 A0A2S2NDF5 A0A2H8U079 A0A0Q9WYB4 B4K8I6 B4PPV3 B4HGC4 B3P196 A0A0A1WIH5 A0A1W4U824 A0A1W4UJ87 B4R1X7 O46036 A0A0K8VGV1 A0A0R1E2T5 A0A0A1WV51 W8AQ12 A0A0R1E2U6 A0A0M4EVQ0 B4JEW9 A0A0K8U655
A0A2W1BDW3 A0A194QC77 A0A1B6F6H4 A0A1B6DLE2 K7INJ3 A0A1B6LTR7 E2ADM8 A0A0N0U6Q9 A0A2J7PI32 V9I9U6 A0A067RB42 A0A0A9YD33 A0A087ZWM2 V9IC45 A0A026WZT5 A0A3L8DRR7 A0A151WRX6 A0A1Y1M9L8 A0A158NPF3 A0A224XC60 A0A310SG02 A0A069DUX2 A0A023F5Q7 A0A0P4VTC3 A0A154PLQ9 A0A0A9XCJ8 A0A0V0G9V3 E0W0A8 U4UHK9 A0A1B6DCA0 N6U4A8 A0A0C9RBU0 A0A1W4WUN9 A0A1B6EC23 A0A1W4X5D5 A0A0A9Y317 A0A0A9Y8M3 A0A0A9Y5K9 A0A1S3CWX9 A0A0A9Y744 A0A0A9Z857 A0A0A9Z291 A0A1S3CVK8 A0A1S3CVR8 V5GVC8 D6X2E0 A0A2A3E232 A0A195E543 A0A1W4WUM5 A0A1W4X5J2 A0A1B0GH93 A0A1I8N265 A0A1I8QDQ6 A0A1I8QDN8 A0A1I8N267 A0A0L0C742 A0A1A9XRG3 A0A1A9UFQ6 A0A1B0BVU9 A0A1A9ZA08 A0A1A9WS45 A0A1B0FIQ7 A0A0T6B5T9 A0A2S2QVF1 A0A1L8E0U4 A0A1L8E0P4 A0A1I8N289 U5ETF2 A0A1I8QDQ7 A0A195BA66 J9JJP6 B4N9Y3 Q299Q6 B4G5K5 A0A2S2NDF5 A0A2H8U079 A0A0Q9WYB4 B4K8I6 B4PPV3 B4HGC4 B3P196 A0A0A1WIH5 A0A1W4U824 A0A1W4UJ87 B4R1X7 O46036 A0A0K8VGV1 A0A0R1E2T5 A0A0A1WV51 W8AQ12 A0A0R1E2U6 A0A0M4EVQ0 B4JEW9 A0A0K8U655
PDB
2HU2
E-value=2.30618e-160,
Score=1452
Ontologies
PATHWAY
GO
GO:0016491
GO:0051287
GO:0016616
GO:0004617
GO:0090090
GO:0003713
GO:0000122
GO:0090263
GO:0006357
GO:0008134
GO:0042802
GO:0035220
GO:0003714
GO:0045944
GO:0042803
GO:0045892
GO:0070491
GO:0016360
GO:0022416
GO:0005634
GO:0043044
GO:0031010
GO:0008152
GO:0008270
GO:0005215
GO:0006270
GO:0042555
GO:0003707
GO:0006281
GO:0006259
GO:0055114
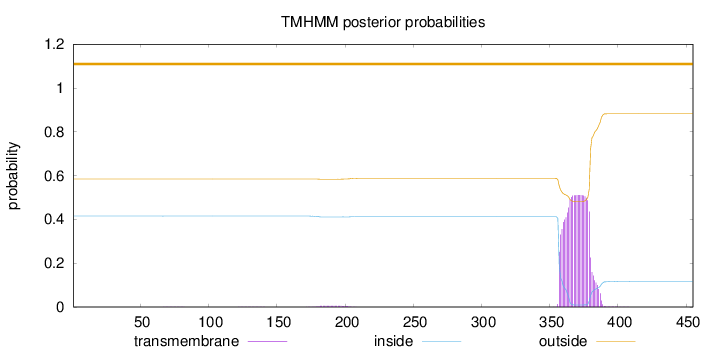
Topology
Subcellular location
Nucleus
Length:
455
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.74667
Exp number, first 60 AAs:
0.00013
Total prob of N-in:
0.41533
outside
1 - 455
Population Genetic Test Statistics
Pi
211.368448
Theta
157.50617
Tajima's D
0.914076
CLR
0.444745
CSRT
0.63666816659167
Interpretation
Uncertain
