Pre Gene Modal
BGIBMGA002230
Annotation
PREDICTED:_macrophage_erythroblast_attacher_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.733
Sequence
CDS
ATGAATGAGATCAAATCTATGGAGCATGCGACATTGAAGGTGCCGTATGAAGTGTTTAACAAGAAGTACCGCAGTGCTCAACGCGTCTTCGATGTTGAGGCGCGTCAGGTTAGCAGTGCCGCCACAGAAATGGAAACGGTCAAGAACCAACCCATCACAAATACAGAGATCGATAGTCTATTGGGTGGCATGGTCGAGAAGTTGACAACGATGAAGCGAAAAGCGAGCGAGGCTATTAACGAAGAGCTCCAGGTCGCTTACGTCTGCAAGAAGCGTCTGGAACACCTGAAGGAGCAGGCCAGCGCCCTCGCCGAGCCGTCCTCCACGCAGACGACGCTGAACCAATGGCGTAAAGTGCGTCTAGATCGGATGCTGGTTGACTACTTCCTGAGGAACGGTTACTACGAGTCCGCTTGGAAGCTGGCGGAATCCGCCGGCCTCAGGGAGCTCACCAATGTCGACATATACAGCGCGGCCGCCGCGGTGGAGGCCGAGCTGAAGAGCCAGAGGACGGCGCGCTGTCTGCAGTGGTGCTCCGGCAACAAGTCCAAGCTGAGGAAGCTGAACTCCACGATGGAGTTCAACATCAGGATACAGGAGTTCATAGAGCTGGTGCGCGAGGACCGGCGCCTGGAGGCCGTGCGTCACGCCAGGAAGCACTTCTCCAACCACGAGGAGGGGCAGCTGGTCGACATTCAGCACTGCATGGGCATGCTGGCCTTCCCCAAGGACACCGAGGTGGAGCCGTACCGGTCGCTGCTGGCGCGCGGGCGCTGGGCGCGGCTGGTGCGGCAGTTCCGCGCCGAGCACGCGCGCCTGCTGCACCCCGCGCCGCTGCCGCTGCTGCCCGCCACGCTGCAGCTGGGCCTGGCCGCGCTCAACACGCCTCAGTGCTACAAGGAGAGCAGCCAGGCGGCGGACTGCCCGGCGTGCCAGGCGCCGCTGTCGGGGCTGGCGAGGCAGCTGCCGCACGCGCACTGCTCGCACTCGCGCCTCGTGTGCCGCATCTCGCGCCTGCCGCTCAACGAGCACAACCAGCCCATGGTGCTGCCCAACGGACAGGTCTACGGGGAGAAGGCTCTGAAGCAAATGATGAAAGAGCACGGCTCGATCATCTGCCCGAAGACGAAAGAAGTGTTTTGCATGAAGCGCGTCGAGAAGGTGTACGTCATGTGA
Protein
MNEIKSMEHATLKVPYEVFNKKYRSAQRVFDVEARQVSSAATEMETVKNQPITNTEIDSLLGGMVEKLTTMKRKASEAINEELQVAYVCKKRLEHLKEQASALAEPSSTQTTLNQWRKVRLDRMLVDYFLRNGYYESAWKLAESAGLRELTNVDIYSAAAAVEAELKSQRTARCLQWCSGNKSKLRKLNSTMEFNIRIQEFIELVREDRRLEAVRHARKHFSNHEEGQLVDIQHCMGMLAFPKDTEVEPYRSLLARGRWARLVRQFRAEHARLLHPAPLPLLPATLQLGLAALNTPQCYKESSQAADCPACQAPLSGLARQLPHAHCSHSRLVCRISRLPLNEHNQPMVLPNGQVYGEKALKQMMKEHGSIICPKTKEVFCMKRVEKVYVM
Summary
Uniprot
H9IY97
A0A2H1W8V7
A0A0N0PAX5
A0A194QCJ8
A0A212ERE5
A0A2W1BCW0
+ More
S4PSB4 A0A067RDP7 A0A1Y1M7W7 A0A2A3E3C1 A0A087ZWL6 A0A0A9YIX7 A0A1E1W327 A0A1B6DDF6 A0A0N0U6Q1 K7INK7 A0A232EU50 A0A2J7PI73 A0A1B6HZ13 A0A0C9QVP5 A0A154PK34 R4WSP4 V5GCP9 A0A0J7P3E8 E9J589 E2ATB5 F4WII5 A0A158NZY7 A0A026WZ02 E2BV49 A0A182ITR2 J9JUX8 B0WGR5 A0A2H8TTD1 A0A084WR10 A0A2R7VNQ3 A0A1B0GH92 A0A139WBG1 A0A1Q3FAE8 A0A182R8F0 A0A182NCM8 A0A182YJM6 A0A182W319 J3JVL4 A0A182M5W8 A0A182TLA1 A0A182LNG7 A0A182WUX8 Q7QAA4 A0A182HVH1 A0A182VLW2 A0A182QNF3 A0A182P3H3 A0A2M3Z6L2 A0A182F8G5 A0A2M4BQS0 U5EUR0 A0A182JYN9 W5JGL4 A0A2M4A651 A0A0V0G838 A0A069DSM1 A0A1L8DD38 E0VTJ6 A0A023ERD2 A0A182SIE8 A0A0N7Z9A5 A0A310S9I6 N6TMD8 U4U8D6 A0A195F2I0 A0A336L2P0 A0A151X3W8 A0A151J0M7 A0A195CLY2 A0A195BMZ5 Q17A38 A0A023FAK6 T1J0Z8 A0A0P4WFY6 A0A1S3HBA0 A0A0K8TQK5 A0A2P6KWP8 A0A182TI57 A0A0C9R9D4 A0A2S2NLA5 V5HE18 B7PXJ7 A0A0L0BN16 T1PB33 R7ULR9 A0A087V0E9 A0A1B0DQC3 A0A1L8EDL2 A0A1I8PW88 B4M0Q6 B3P6X9 B4QTT4 B4PV36 B4IJC6
S4PSB4 A0A067RDP7 A0A1Y1M7W7 A0A2A3E3C1 A0A087ZWL6 A0A0A9YIX7 A0A1E1W327 A0A1B6DDF6 A0A0N0U6Q1 K7INK7 A0A232EU50 A0A2J7PI73 A0A1B6HZ13 A0A0C9QVP5 A0A154PK34 R4WSP4 V5GCP9 A0A0J7P3E8 E9J589 E2ATB5 F4WII5 A0A158NZY7 A0A026WZ02 E2BV49 A0A182ITR2 J9JUX8 B0WGR5 A0A2H8TTD1 A0A084WR10 A0A2R7VNQ3 A0A1B0GH92 A0A139WBG1 A0A1Q3FAE8 A0A182R8F0 A0A182NCM8 A0A182YJM6 A0A182W319 J3JVL4 A0A182M5W8 A0A182TLA1 A0A182LNG7 A0A182WUX8 Q7QAA4 A0A182HVH1 A0A182VLW2 A0A182QNF3 A0A182P3H3 A0A2M3Z6L2 A0A182F8G5 A0A2M4BQS0 U5EUR0 A0A182JYN9 W5JGL4 A0A2M4A651 A0A0V0G838 A0A069DSM1 A0A1L8DD38 E0VTJ6 A0A023ERD2 A0A182SIE8 A0A0N7Z9A5 A0A310S9I6 N6TMD8 U4U8D6 A0A195F2I0 A0A336L2P0 A0A151X3W8 A0A151J0M7 A0A195CLY2 A0A195BMZ5 Q17A38 A0A023FAK6 T1J0Z8 A0A0P4WFY6 A0A1S3HBA0 A0A0K8TQK5 A0A2P6KWP8 A0A182TI57 A0A0C9R9D4 A0A2S2NLA5 V5HE18 B7PXJ7 A0A0L0BN16 T1PB33 R7ULR9 A0A087V0E9 A0A1B0DQC3 A0A1L8EDL2 A0A1I8PW88 B4M0Q6 B3P6X9 B4QTT4 B4PV36 B4IJC6
Pubmed
19121390
26354079
22118469
28756777
23622113
24845553
+ More
28004739 25401762 26823975 20075255 28648823 23691247 21282665 20798317 21719571 21347285 24508170 30249741 24438588 18362917 19820115 25244985 22516182 20966253 12364791 14747013 17210077 20920257 23761445 26334808 20566863 24945155 27129103 23537049 17510324 25474469 26369729 25765539 26108605 25315136 23254933 17994087 17550304
28004739 25401762 26823975 20075255 28648823 23691247 21282665 20798317 21719571 21347285 24508170 30249741 24438588 18362917 19820115 25244985 22516182 20966253 12364791 14747013 17210077 20920257 23761445 26334808 20566863 24945155 27129103 23537049 17510324 25474469 26369729 25765539 26108605 25315136 23254933 17994087 17550304
EMBL
BABH01020900
BABH01020901
ODYU01006953
SOQ49272.1
KQ461188
KPJ07321.1
+ More
KQ459193 KPJ03144.1 AGBW02013027 OWR44068.1 KZ150129 PZC73079.1 GAIX01014003 JAA78557.1 KK852766 KDR16975.1 GEZM01037866 JAV81872.1 KZ288438 PBC25746.1 GBHO01010567 GBRD01008606 GDHC01002620 JAG33037.1 JAG57215.1 JAQ16009.1 GDQN01009684 JAT81370.1 GEDC01013589 JAS23709.1 KQ435721 KOX78524.1 NNAY01002170 OXU21883.1 NEVH01025129 PNF16028.1 GECU01027790 JAS79916.1 GBYB01004737 JAG74504.1 KQ434938 KZC12157.1 AK417732 BAN20947.1 GALX01000556 JAB67910.1 LBMM01000214 KMR04459.1 GL768124 EFZ12028.1 GL442548 EFN63282.1 GL888175 EGI66116.1 ADTU01005133 KK107063 QOIP01000005 EZA61257.1 RLU22639.1 GL450810 EFN80439.1 ABLF02024977 ABLF02024981 DS231928 EDS27141.1 GFXV01004653 MBW16458.1 ATLV01025875 KE525402 KFB52654.1 KK854007 PTY09167.1 AJWK01004085 KQ971372 KYB25256.1 GFDL01010553 JAV24492.1 BT127282 AEE62244.1 AXCM01000300 AAAB01008898 EAA09156.2 APCN01005777 AXCN02001183 GGFM01003415 MBW24166.1 GGFJ01006182 MBW55323.1 GANO01001357 JAB58514.1 ADMH02001252 ETN63502.1 GGFK01002889 MBW36210.1 GECL01002067 JAP04057.1 GBGD01001801 JAC87088.1 GFDF01009844 JAV04240.1 DS235767 EEB16723.1 GAPW01001858 JAC11740.1 GDKW01001091 JAI55504.1 KQ768431 OAD53171.1 APGK01018646 KB740081 ENN81629.1 KB632220 ERL90184.1 KQ981856 KYN34591.1 UFQS01000883 UFQT01000883 SSX07480.1 SSX27820.1 KQ982557 KYQ55107.1 KQ980624 KYN15034.1 KQ977580 KYN01718.1 KQ976440 KYM86554.1 CH477340 EAT43126.1 GBBI01000220 JAC18492.1 JH431742 GDRN01060114 JAI65429.1 GDAI01000956 JAI16647.1 MWRG01004217 PRD30732.1 GBYB01004735 JAG74502.1 GGMR01004937 MBY17556.1 GANP01002608 JAB81860.1 ABJB010800282 DS814574 EEC11319.1 JRES01001708 KNC20674.1 KA645904 AFP60533.1 AMQN01007212 KB300220 ELU07028.1 KK122591 KFM83088.1 AJVK01019041 GFDG01002100 JAV16699.1 CH940650 EDW67348.1 CH954182 EDV53799.1 CM000364 EDX14288.1 CM000160 EDW98885.1 CH480848 EDW51117.1
KQ459193 KPJ03144.1 AGBW02013027 OWR44068.1 KZ150129 PZC73079.1 GAIX01014003 JAA78557.1 KK852766 KDR16975.1 GEZM01037866 JAV81872.1 KZ288438 PBC25746.1 GBHO01010567 GBRD01008606 GDHC01002620 JAG33037.1 JAG57215.1 JAQ16009.1 GDQN01009684 JAT81370.1 GEDC01013589 JAS23709.1 KQ435721 KOX78524.1 NNAY01002170 OXU21883.1 NEVH01025129 PNF16028.1 GECU01027790 JAS79916.1 GBYB01004737 JAG74504.1 KQ434938 KZC12157.1 AK417732 BAN20947.1 GALX01000556 JAB67910.1 LBMM01000214 KMR04459.1 GL768124 EFZ12028.1 GL442548 EFN63282.1 GL888175 EGI66116.1 ADTU01005133 KK107063 QOIP01000005 EZA61257.1 RLU22639.1 GL450810 EFN80439.1 ABLF02024977 ABLF02024981 DS231928 EDS27141.1 GFXV01004653 MBW16458.1 ATLV01025875 KE525402 KFB52654.1 KK854007 PTY09167.1 AJWK01004085 KQ971372 KYB25256.1 GFDL01010553 JAV24492.1 BT127282 AEE62244.1 AXCM01000300 AAAB01008898 EAA09156.2 APCN01005777 AXCN02001183 GGFM01003415 MBW24166.1 GGFJ01006182 MBW55323.1 GANO01001357 JAB58514.1 ADMH02001252 ETN63502.1 GGFK01002889 MBW36210.1 GECL01002067 JAP04057.1 GBGD01001801 JAC87088.1 GFDF01009844 JAV04240.1 DS235767 EEB16723.1 GAPW01001858 JAC11740.1 GDKW01001091 JAI55504.1 KQ768431 OAD53171.1 APGK01018646 KB740081 ENN81629.1 KB632220 ERL90184.1 KQ981856 KYN34591.1 UFQS01000883 UFQT01000883 SSX07480.1 SSX27820.1 KQ982557 KYQ55107.1 KQ980624 KYN15034.1 KQ977580 KYN01718.1 KQ976440 KYM86554.1 CH477340 EAT43126.1 GBBI01000220 JAC18492.1 JH431742 GDRN01060114 JAI65429.1 GDAI01000956 JAI16647.1 MWRG01004217 PRD30732.1 GBYB01004735 JAG74502.1 GGMR01004937 MBY17556.1 GANP01002608 JAB81860.1 ABJB010800282 DS814574 EEC11319.1 JRES01001708 KNC20674.1 KA645904 AFP60533.1 AMQN01007212 KB300220 ELU07028.1 KK122591 KFM83088.1 AJVK01019041 GFDG01002100 JAV16699.1 CH940650 EDW67348.1 CH954182 EDV53799.1 CM000364 EDX14288.1 CM000160 EDW98885.1 CH480848 EDW51117.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000027135
UP000242457
+ More
UP000005203 UP000053105 UP000002358 UP000215335 UP000235965 UP000076502 UP000036403 UP000000311 UP000007755 UP000005205 UP000053097 UP000279307 UP000008237 UP000075880 UP000007819 UP000002320 UP000030765 UP000092461 UP000007266 UP000075900 UP000075884 UP000076408 UP000075920 UP000075883 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075903 UP000075886 UP000075885 UP000069272 UP000075881 UP000000673 UP000009046 UP000075901 UP000019118 UP000030742 UP000078541 UP000075809 UP000078492 UP000078542 UP000078540 UP000008820 UP000085678 UP000001555 UP000037069 UP000095301 UP000014760 UP000054359 UP000092462 UP000095300 UP000008792 UP000008711 UP000000304 UP000002282 UP000001292
UP000005203 UP000053105 UP000002358 UP000215335 UP000235965 UP000076502 UP000036403 UP000000311 UP000007755 UP000005205 UP000053097 UP000279307 UP000008237 UP000075880 UP000007819 UP000002320 UP000030765 UP000092461 UP000007266 UP000075900 UP000075884 UP000076408 UP000075920 UP000075883 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000075903 UP000075886 UP000075885 UP000069272 UP000075881 UP000000673 UP000009046 UP000075901 UP000019118 UP000030742 UP000078541 UP000075809 UP000078492 UP000078542 UP000078540 UP000008820 UP000085678 UP000001555 UP000037069 UP000095301 UP000014760 UP000054359 UP000092462 UP000095300 UP000008792 UP000008711 UP000000304 UP000002282 UP000001292
PRIDE
Pfam
PF10607 CLTH
Interpro
Gene 3D
ProteinModelPortal
H9IY97
A0A2H1W8V7
A0A0N0PAX5
A0A194QCJ8
A0A212ERE5
A0A2W1BCW0
+ More
S4PSB4 A0A067RDP7 A0A1Y1M7W7 A0A2A3E3C1 A0A087ZWL6 A0A0A9YIX7 A0A1E1W327 A0A1B6DDF6 A0A0N0U6Q1 K7INK7 A0A232EU50 A0A2J7PI73 A0A1B6HZ13 A0A0C9QVP5 A0A154PK34 R4WSP4 V5GCP9 A0A0J7P3E8 E9J589 E2ATB5 F4WII5 A0A158NZY7 A0A026WZ02 E2BV49 A0A182ITR2 J9JUX8 B0WGR5 A0A2H8TTD1 A0A084WR10 A0A2R7VNQ3 A0A1B0GH92 A0A139WBG1 A0A1Q3FAE8 A0A182R8F0 A0A182NCM8 A0A182YJM6 A0A182W319 J3JVL4 A0A182M5W8 A0A182TLA1 A0A182LNG7 A0A182WUX8 Q7QAA4 A0A182HVH1 A0A182VLW2 A0A182QNF3 A0A182P3H3 A0A2M3Z6L2 A0A182F8G5 A0A2M4BQS0 U5EUR0 A0A182JYN9 W5JGL4 A0A2M4A651 A0A0V0G838 A0A069DSM1 A0A1L8DD38 E0VTJ6 A0A023ERD2 A0A182SIE8 A0A0N7Z9A5 A0A310S9I6 N6TMD8 U4U8D6 A0A195F2I0 A0A336L2P0 A0A151X3W8 A0A151J0M7 A0A195CLY2 A0A195BMZ5 Q17A38 A0A023FAK6 T1J0Z8 A0A0P4WFY6 A0A1S3HBA0 A0A0K8TQK5 A0A2P6KWP8 A0A182TI57 A0A0C9R9D4 A0A2S2NLA5 V5HE18 B7PXJ7 A0A0L0BN16 T1PB33 R7ULR9 A0A087V0E9 A0A1B0DQC3 A0A1L8EDL2 A0A1I8PW88 B4M0Q6 B3P6X9 B4QTT4 B4PV36 B4IJC6
S4PSB4 A0A067RDP7 A0A1Y1M7W7 A0A2A3E3C1 A0A087ZWL6 A0A0A9YIX7 A0A1E1W327 A0A1B6DDF6 A0A0N0U6Q1 K7INK7 A0A232EU50 A0A2J7PI73 A0A1B6HZ13 A0A0C9QVP5 A0A154PK34 R4WSP4 V5GCP9 A0A0J7P3E8 E9J589 E2ATB5 F4WII5 A0A158NZY7 A0A026WZ02 E2BV49 A0A182ITR2 J9JUX8 B0WGR5 A0A2H8TTD1 A0A084WR10 A0A2R7VNQ3 A0A1B0GH92 A0A139WBG1 A0A1Q3FAE8 A0A182R8F0 A0A182NCM8 A0A182YJM6 A0A182W319 J3JVL4 A0A182M5W8 A0A182TLA1 A0A182LNG7 A0A182WUX8 Q7QAA4 A0A182HVH1 A0A182VLW2 A0A182QNF3 A0A182P3H3 A0A2M3Z6L2 A0A182F8G5 A0A2M4BQS0 U5EUR0 A0A182JYN9 W5JGL4 A0A2M4A651 A0A0V0G838 A0A069DSM1 A0A1L8DD38 E0VTJ6 A0A023ERD2 A0A182SIE8 A0A0N7Z9A5 A0A310S9I6 N6TMD8 U4U8D6 A0A195F2I0 A0A336L2P0 A0A151X3W8 A0A151J0M7 A0A195CLY2 A0A195BMZ5 Q17A38 A0A023FAK6 T1J0Z8 A0A0P4WFY6 A0A1S3HBA0 A0A0K8TQK5 A0A2P6KWP8 A0A182TI57 A0A0C9R9D4 A0A2S2NLA5 V5HE18 B7PXJ7 A0A0L0BN16 T1PB33 R7ULR9 A0A087V0E9 A0A1B0DQC3 A0A1L8EDL2 A0A1I8PW88 B4M0Q6 B3P6X9 B4QTT4 B4PV36 B4IJC6
Ontologies
GO
PANTHER
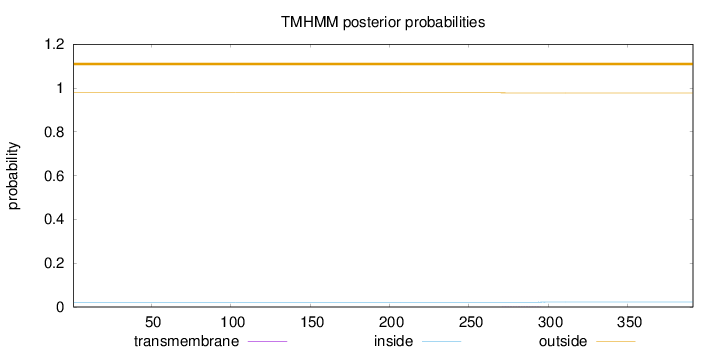
Topology
Length:
391
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02646
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02182
outside
1 - 391
Population Genetic Test Statistics
Pi
253.462276
Theta
179.333328
Tajima's D
1.293244
CLR
0.074111
CSRT
0.741962901854907
Interpretation
Uncertain
