Gene
KWMTBOMO15779
Pre Gene Modal
BGIBMGA002345
Annotation
PREDICTED:_zinc_transporter_ZIP13_homolog_[Papilio_xuthus]
Full name
Zinc transporter ZIP13 homolog
+ More
Mediator of RNA polymerase II transcription subunit 15
Mediator of RNA polymerase II transcription subunit 15
Alternative Name
Zinc/iron regulated transporter-related protein 99C
Mediator complex subunit 15
Mediator complex subunit 15
Location in the cell
PlasmaMembrane Reliability : 3.116
Sequence
CDS
ATGGCGAGCTACGTGCCAGAATATTTGTCCAATTTTGTGGAAGAATTAGATGAACATCCGTGGATTTTCGCTTCGCTGGCGTCGATGCTGGTCGGTTTAAGCGGCATCTTACCGCTGTTCATCATTCCACTGGATTCTTCAGCCACATTCGAAGATGGGCCCGGGGCAGCCACACTTCGGATCCTGCTCAGTTTCGCTGTAGGCGGTCTACTGGGCGATGTCTTCCTCCACCTACTACCGGAAGCGTGGGAAAATGATATGGCCAACACTAAAGCGGGACAACACGTTTCTCTCAAGAGCGGTCTGTGGTGTCTGTTCGGTATGCTGGTCTTCATAGTCGTGGAGAAGCTGTTCTCGGTGGACGAGCACAACGACGGCGAAGAGGAGGAGGAGAAGCAGAACGGCGCAATAAAGCAGATAGAGATCGAAGAGATCGAGAAACTGCTCGAGATCGAAAGGAAACATCGGGGAGTGGATAAGGGGATCTGCAATGGAACGGTGACCGCCAAGCAGATATACGATTCTTGTATCTTCAATAATAACACTAAAGGCGAAGGAGGATGTTGCAGTATGCCTACAAGCAACGCTGGAGCCAGCGGCCGCTGTAAGGGCTCCTCTCGCTGGATGGGGCGCTGCTTGCTTCGGGAAGCTCGCGAGAAAGCACTCCGCGCCAGTAAAGAGAAAACTGAGAAGAAAGATGTGGCGGGCTATTTGAATCTGATGGCGAACTCGATCGACAACTTCACCCACGGACTCGCGGTCGGGGGCTCGTTCCTCGTCGGGTTCCGGGTCGGAGTTCTCACCACCTTTGCCATATTAGTGCACGAGATCCCGCACGAGGTGGGAGACTTCGCCATACTGCTGAAGAGCGGGTTCTCCAGGTGGGAGGCCGCCAAAGCACAACTGGCCACGGCGACGGCGGGGCTGGTGGGCGCCATGACGGCCGTCGTGTTCAGCGGCGCCAGCAATTCGCTCGAAGCGAAGACCTCGTGGATCGTGCCGTTCACGGCGGGCGGGTTCCTGCACATCGCGCTGGTGACGGTGCTGCCGGACCTGCTGCGGGAGCACTCCAAGCTGGAGTCCCTAAAGCACTTCGCGGCGCTCGCCCTCGGCATCCTGCTCATGCACACCCTCACCACAGTTCATAGAGAGATGTAA
Protein
MASYVPEYLSNFVEELDEHPWIFASLASMLVGLSGILPLFIIPLDSSATFEDGPGAATLRILLSFAVGGLLGDVFLHLLPEAWENDMANTKAGQHVSLKSGLWCLFGMLVFIVVEKLFSVDEHNDGEEEEEKQNGAIKQIEIEEIEKLLEIERKHRGVDKGICNGTVTAKQIYDSCIFNNNTKGEGGCCSMPTSNAGASGRCKGSSRWMGRCLLREAREKALRASKEKTEKKDVAGYLNLMANSIDNFTHGLAVGGSFLVGFRVGVLTTFAILVHEIPHEVGDFAILLKSGFSRWEAAKAQLATATAGLVGAMTAVVFSGASNSLEAKTSWIVPFTAGGFLHIALVTVLPDLLREHSKLESLKHFAALALGILLMHTLTTVHREM
Summary
Description
Involved in zinc transport and homeostasis.
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors.
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors.
Subunit
Component of the Mediator complex.
Similarity
Belongs to the ZIP transporter (TC 2.A.5) family. KE4/Catsup subfamily.
Belongs to the Mediator complex subunit 15 family.
Belongs to the Mediator complex subunit 15 family.
Keywords
Cell membrane
Complete proteome
Glycoprotein
Golgi apparatus
Ion transport
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Transport
Zinc
Zinc transport
Feature
chain Zinc transporter ZIP13 homolog
Uniprot
H9IYL1
A0A0N1PFS4
A0A194QDW8
A0A2W1BK27
A0A212ERG0
A0A1A9W7Y1
+ More
A0A0A1WY74 A0A0A1XNR0 A0A0M5JCT1 A0A0K8TLC1 B4R0U7 B4HZG9 B3LYT7 A4V3L8 Q9VAF0 B4NK13 A0A034WJV5 A0A1B0AYC9 D3TSB2 A0A0L0C7J4 B4M5R5 A0A1W4URN3 B4PPI2 B4K5Z6 A0A0K8W0P3 A0A3B0JFR1 A0A0K8VS14 B4JYL0 B4GNJ6 Q29C60 D0QWQ1 B3P5B3 A0A0R3NF67 A0A1A9Z8A5 Q16FG6 A0A1A9XI93 A0A1A9V3F4 W8BM22 A0A2M4AGL3 A0A2M4AIJ7 A0A2M3ZB96 A0A1L8DEG1 A0A1L8DEH9 A0A1L8DE02 U5EX14 W5JIU1 A0A1J1IVX9 K7IR27 A0A182MM77 A0A1B0CR50 A0A1I8NNW4 A0A336LUY9 A0A232F907 A0A1L8EDW2 T1PKA0 A0A182KDQ8 A0A1L8DDI0 A0A1B6E318 A0A1Q3G3M6 A0A182RHY3 A0A182FNG5 A0A2M4BP99 A0A2M4BQ56 A0A1B6J1S2 A0A1B0FB02 T1DTA1 A0A182VQL6 A0A182P5R3 A0A182QC95 A0A182V8M6 A0A182IIW3 A0A1B6FC85 A0A182JAY0 A0A182LCR9 Q7PLY3 A0A2S2PTK2 A0A182NL67 A0A023ENP7 A0A1B0DEG7 A0A195B1U7 A0A2H8TYI2 A0A182X1Z0 A0A1L8E8X5 A0A151JQA1 A0A1Q3FF19 A0A2J7PUE6 A0A1S3IPE5 A0A182TLS0 A0A3M6U720 C3Y490 A0A3B3R623 A0A3Q1G139 A0A3Q3KWJ0 A0A3Q3R3B1 A0A3B4TYZ5 A0A3P8ZTA2 A0A3Q3KK36 A0A3B4YCT0 A0A3Q1HZQ2
A0A0A1WY74 A0A0A1XNR0 A0A0M5JCT1 A0A0K8TLC1 B4R0U7 B4HZG9 B3LYT7 A4V3L8 Q9VAF0 B4NK13 A0A034WJV5 A0A1B0AYC9 D3TSB2 A0A0L0C7J4 B4M5R5 A0A1W4URN3 B4PPI2 B4K5Z6 A0A0K8W0P3 A0A3B0JFR1 A0A0K8VS14 B4JYL0 B4GNJ6 Q29C60 D0QWQ1 B3P5B3 A0A0R3NF67 A0A1A9Z8A5 Q16FG6 A0A1A9XI93 A0A1A9V3F4 W8BM22 A0A2M4AGL3 A0A2M4AIJ7 A0A2M3ZB96 A0A1L8DEG1 A0A1L8DEH9 A0A1L8DE02 U5EX14 W5JIU1 A0A1J1IVX9 K7IR27 A0A182MM77 A0A1B0CR50 A0A1I8NNW4 A0A336LUY9 A0A232F907 A0A1L8EDW2 T1PKA0 A0A182KDQ8 A0A1L8DDI0 A0A1B6E318 A0A1Q3G3M6 A0A182RHY3 A0A182FNG5 A0A2M4BP99 A0A2M4BQ56 A0A1B6J1S2 A0A1B0FB02 T1DTA1 A0A182VQL6 A0A182P5R3 A0A182QC95 A0A182V8M6 A0A182IIW3 A0A1B6FC85 A0A182JAY0 A0A182LCR9 Q7PLY3 A0A2S2PTK2 A0A182NL67 A0A023ENP7 A0A1B0DEG7 A0A195B1U7 A0A2H8TYI2 A0A182X1Z0 A0A1L8E8X5 A0A151JQA1 A0A1Q3FF19 A0A2J7PUE6 A0A1S3IPE5 A0A182TLS0 A0A3M6U720 C3Y490 A0A3B3R623 A0A3Q1G139 A0A3Q3KWJ0 A0A3Q3R3B1 A0A3B4TYZ5 A0A3P8ZTA2 A0A3Q3KK36 A0A3B4YCT0 A0A3Q1HZQ2
Pubmed
19121390
26354079
28756777
22118469
25830018
26369729
+ More
17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 23322169 25348373 20353571 26108605 18057021 17550304 15632085 23185243 19859648 17510324 24495485 20920257 23761445 20075255 28648823 25315136 20966253 12364791 14747013 17210077 24945155 30382153 18563158 29240929 25069045
17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 23322169 25348373 20353571 26108605 18057021 17550304 15632085 23185243 19859648 17510324 24495485 20920257 23761445 20075255 28648823 25315136 20966253 12364791 14747013 17210077 24945155 30382153 18563158 29240929 25069045
EMBL
BABH01020899
KQ461188
KPJ07322.1
KQ459193
KPJ03145.1
KZ150129
+ More
PZC73080.1 AGBW02013027 OWR44067.1 GBXI01014723 GBXI01010964 JAC99568.1 JAD03328.1 GBXI01014111 GBXI01010992 GBXI01001328 JAD00181.1 JAD03300.1 JAD12964.1 CP012526 ALC47168.1 GDAI01002888 JAI14715.1 CM000364 EDX14918.1 CH480819 EDW53426.1 CH902617 EDV44053.1 AE014297 AAN14196.1 AAN14197.1 AAN14198.1 AAN14199.1 ADV37403.1 ADV37404.1 ADV37405.1 BT001774 CH964272 EDW84015.1 GAKP01004001 GAKP01004000 GAKP01003999 GAKP01003998 JAC54954.1 JXJN01005666 JXJN01005667 JXJN01005668 EZ424314 ADD20590.1 JRES01000809 KNC28221.1 CH940652 EDW58991.1 KRF78632.1 CM000160 EDW98232.1 KRK04169.1 CH933806 EDW16233.1 GDHF01016177 GDHF01007648 JAI36137.1 JAI44666.1 OUUW01000005 SPP80965.1 GDHF01010964 JAI41350.1 CH916377 EDV90772.1 CH479186 EDW39322.1 CM000070 EAL26786.1 EIM52297.1 KRS99548.1 KRS99549.1 FJ821034 ACN94762.1 CH954182 EDV53163.1 KRS99547.1 CH478442 EAT32983.1 GAMC01012136 GAMC01012134 JAB94421.1 GGFK01006595 MBW39916.1 GGFK01007294 MBW40615.1 GGFM01005020 MBW25771.1 GFDF01009232 JAV04852.1 GFDF01009233 JAV04851.1 GFDF01009540 JAV04544.1 GANO01000295 JAB59576.1 ADMH02001124 ETN64041.1 CVRI01000058 CRL02697.1 AXCM01004511 AJWK01024397 UFQT01000202 SSX21660.1 NNAY01000669 OXU27082.1 GFDG01002020 JAV16779.1 KA649190 KA649191 AFP63820.1 GFDF01009555 JAV04529.1 GEDC01005011 JAS32287.1 GFDL01000679 JAV34366.1 GGFJ01005756 MBW54897.1 GGFJ01005757 MBW54898.1 GECU01014600 GECU01014553 JAS93106.1 JAS93153.1 CCAG010023703 GAMD01001094 JAB00497.1 AXCN02000083 APCN01002160 GECZ01022206 GECZ01006067 JAS47563.1 JAS63702.1 AAAB01008982 EAA14648.3 GGMR01020088 MBY32707.1 GAPW01003038 JAC10560.1 AJVK01032733 AJVK01032734 AJVK01032735 KQ976667 KYM78260.1 GFXV01007046 MBW18851.1 GFDG01003767 JAV15032.1 KQ978700 KYN29147.1 GFDL01008906 JAV26139.1 NEVH01021199 PNF19953.1 RCHS01002149 RMX49339.1 GG666484 EEN65116.1
PZC73080.1 AGBW02013027 OWR44067.1 GBXI01014723 GBXI01010964 JAC99568.1 JAD03328.1 GBXI01014111 GBXI01010992 GBXI01001328 JAD00181.1 JAD03300.1 JAD12964.1 CP012526 ALC47168.1 GDAI01002888 JAI14715.1 CM000364 EDX14918.1 CH480819 EDW53426.1 CH902617 EDV44053.1 AE014297 AAN14196.1 AAN14197.1 AAN14198.1 AAN14199.1 ADV37403.1 ADV37404.1 ADV37405.1 BT001774 CH964272 EDW84015.1 GAKP01004001 GAKP01004000 GAKP01003999 GAKP01003998 JAC54954.1 JXJN01005666 JXJN01005667 JXJN01005668 EZ424314 ADD20590.1 JRES01000809 KNC28221.1 CH940652 EDW58991.1 KRF78632.1 CM000160 EDW98232.1 KRK04169.1 CH933806 EDW16233.1 GDHF01016177 GDHF01007648 JAI36137.1 JAI44666.1 OUUW01000005 SPP80965.1 GDHF01010964 JAI41350.1 CH916377 EDV90772.1 CH479186 EDW39322.1 CM000070 EAL26786.1 EIM52297.1 KRS99548.1 KRS99549.1 FJ821034 ACN94762.1 CH954182 EDV53163.1 KRS99547.1 CH478442 EAT32983.1 GAMC01012136 GAMC01012134 JAB94421.1 GGFK01006595 MBW39916.1 GGFK01007294 MBW40615.1 GGFM01005020 MBW25771.1 GFDF01009232 JAV04852.1 GFDF01009233 JAV04851.1 GFDF01009540 JAV04544.1 GANO01000295 JAB59576.1 ADMH02001124 ETN64041.1 CVRI01000058 CRL02697.1 AXCM01004511 AJWK01024397 UFQT01000202 SSX21660.1 NNAY01000669 OXU27082.1 GFDG01002020 JAV16779.1 KA649190 KA649191 AFP63820.1 GFDF01009555 JAV04529.1 GEDC01005011 JAS32287.1 GFDL01000679 JAV34366.1 GGFJ01005756 MBW54897.1 GGFJ01005757 MBW54898.1 GECU01014600 GECU01014553 JAS93106.1 JAS93153.1 CCAG010023703 GAMD01001094 JAB00497.1 AXCN02000083 APCN01002160 GECZ01022206 GECZ01006067 JAS47563.1 JAS63702.1 AAAB01008982 EAA14648.3 GGMR01020088 MBY32707.1 GAPW01003038 JAC10560.1 AJVK01032733 AJVK01032734 AJVK01032735 KQ976667 KYM78260.1 GFXV01007046 MBW18851.1 GFDG01003767 JAV15032.1 KQ978700 KYN29147.1 GFDL01008906 JAV26139.1 NEVH01021199 PNF19953.1 RCHS01002149 RMX49339.1 GG666484 EEN65116.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000091820
UP000092553
+ More
UP000000304 UP000001292 UP000007801 UP000000803 UP000007798 UP000092460 UP000037069 UP000008792 UP000192221 UP000002282 UP000009192 UP000268350 UP000001070 UP000008744 UP000001819 UP000008711 UP000092445 UP000008820 UP000092443 UP000078200 UP000000673 UP000183832 UP000002358 UP000075883 UP000092461 UP000095300 UP000215335 UP000095301 UP000075881 UP000075900 UP000069272 UP000092444 UP000075920 UP000075885 UP000075886 UP000075903 UP000075840 UP000075880 UP000075882 UP000007062 UP000075884 UP000092462 UP000078540 UP000076407 UP000078492 UP000235965 UP000085678 UP000075902 UP000275408 UP000001554 UP000261540 UP000257200 UP000261640 UP000261600 UP000261420 UP000265140 UP000261360 UP000265040
UP000000304 UP000001292 UP000007801 UP000000803 UP000007798 UP000092460 UP000037069 UP000008792 UP000192221 UP000002282 UP000009192 UP000268350 UP000001070 UP000008744 UP000001819 UP000008711 UP000092445 UP000008820 UP000092443 UP000078200 UP000000673 UP000183832 UP000002358 UP000075883 UP000092461 UP000095300 UP000215335 UP000095301 UP000075881 UP000075900 UP000069272 UP000092444 UP000075920 UP000075885 UP000075886 UP000075903 UP000075840 UP000075880 UP000075882 UP000007062 UP000075884 UP000092462 UP000078540 UP000076407 UP000078492 UP000235965 UP000085678 UP000075902 UP000275408 UP000001554 UP000261540 UP000257200 UP000261640 UP000261600 UP000261420 UP000265140 UP000261360 UP000265040
ProteinModelPortal
H9IYL1
A0A0N1PFS4
A0A194QDW8
A0A2W1BK27
A0A212ERG0
A0A1A9W7Y1
+ More
A0A0A1WY74 A0A0A1XNR0 A0A0M5JCT1 A0A0K8TLC1 B4R0U7 B4HZG9 B3LYT7 A4V3L8 Q9VAF0 B4NK13 A0A034WJV5 A0A1B0AYC9 D3TSB2 A0A0L0C7J4 B4M5R5 A0A1W4URN3 B4PPI2 B4K5Z6 A0A0K8W0P3 A0A3B0JFR1 A0A0K8VS14 B4JYL0 B4GNJ6 Q29C60 D0QWQ1 B3P5B3 A0A0R3NF67 A0A1A9Z8A5 Q16FG6 A0A1A9XI93 A0A1A9V3F4 W8BM22 A0A2M4AGL3 A0A2M4AIJ7 A0A2M3ZB96 A0A1L8DEG1 A0A1L8DEH9 A0A1L8DE02 U5EX14 W5JIU1 A0A1J1IVX9 K7IR27 A0A182MM77 A0A1B0CR50 A0A1I8NNW4 A0A336LUY9 A0A232F907 A0A1L8EDW2 T1PKA0 A0A182KDQ8 A0A1L8DDI0 A0A1B6E318 A0A1Q3G3M6 A0A182RHY3 A0A182FNG5 A0A2M4BP99 A0A2M4BQ56 A0A1B6J1S2 A0A1B0FB02 T1DTA1 A0A182VQL6 A0A182P5R3 A0A182QC95 A0A182V8M6 A0A182IIW3 A0A1B6FC85 A0A182JAY0 A0A182LCR9 Q7PLY3 A0A2S2PTK2 A0A182NL67 A0A023ENP7 A0A1B0DEG7 A0A195B1U7 A0A2H8TYI2 A0A182X1Z0 A0A1L8E8X5 A0A151JQA1 A0A1Q3FF19 A0A2J7PUE6 A0A1S3IPE5 A0A182TLS0 A0A3M6U720 C3Y490 A0A3B3R623 A0A3Q1G139 A0A3Q3KWJ0 A0A3Q3R3B1 A0A3B4TYZ5 A0A3P8ZTA2 A0A3Q3KK36 A0A3B4YCT0 A0A3Q1HZQ2
A0A0A1WY74 A0A0A1XNR0 A0A0M5JCT1 A0A0K8TLC1 B4R0U7 B4HZG9 B3LYT7 A4V3L8 Q9VAF0 B4NK13 A0A034WJV5 A0A1B0AYC9 D3TSB2 A0A0L0C7J4 B4M5R5 A0A1W4URN3 B4PPI2 B4K5Z6 A0A0K8W0P3 A0A3B0JFR1 A0A0K8VS14 B4JYL0 B4GNJ6 Q29C60 D0QWQ1 B3P5B3 A0A0R3NF67 A0A1A9Z8A5 Q16FG6 A0A1A9XI93 A0A1A9V3F4 W8BM22 A0A2M4AGL3 A0A2M4AIJ7 A0A2M3ZB96 A0A1L8DEG1 A0A1L8DEH9 A0A1L8DE02 U5EX14 W5JIU1 A0A1J1IVX9 K7IR27 A0A182MM77 A0A1B0CR50 A0A1I8NNW4 A0A336LUY9 A0A232F907 A0A1L8EDW2 T1PKA0 A0A182KDQ8 A0A1L8DDI0 A0A1B6E318 A0A1Q3G3M6 A0A182RHY3 A0A182FNG5 A0A2M4BP99 A0A2M4BQ56 A0A1B6J1S2 A0A1B0FB02 T1DTA1 A0A182VQL6 A0A182P5R3 A0A182QC95 A0A182V8M6 A0A182IIW3 A0A1B6FC85 A0A182JAY0 A0A182LCR9 Q7PLY3 A0A2S2PTK2 A0A182NL67 A0A023ENP7 A0A1B0DEG7 A0A195B1U7 A0A2H8TYI2 A0A182X1Z0 A0A1L8E8X5 A0A151JQA1 A0A1Q3FF19 A0A2J7PUE6 A0A1S3IPE5 A0A182TLS0 A0A3M6U720 C3Y490 A0A3B3R623 A0A3Q1G139 A0A3Q3KWJ0 A0A3Q3R3B1 A0A3B4TYZ5 A0A3P8ZTA2 A0A3Q3KK36 A0A3B4YCT0 A0A3Q1HZQ2
Ontologies
GO
GO:0046873
GO:0016021
GO:0016323
GO:0005794
GO:0010312
GO:0060586
GO:0005381
GO:0005783
GO:0005385
GO:0006826
GO:0000139
GO:0016592
GO:0006357
GO:0003712
GO:0008430
GO:0005623
GO:0006882
GO:0071577
GO:0016020
GO:0030001
GO:0008270
GO:0005215
GO:0006270
GO:0042555
GO:0003707
GO:0006281
GO:0006259
GO:0055085
PANTHER
Topology
Subcellular location
Basolateral cell membrane
Golgi apparatus membrane
Nucleus
Golgi apparatus membrane
Nucleus
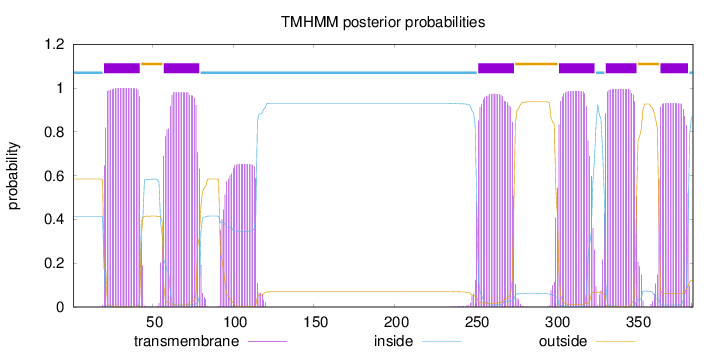
Length:
385
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
140.16663
Exp number, first 60 AAs:
26.48587
Total prob of N-in:
0.41503
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 42
outside
43 - 56
TMhelix
57 - 79
inside
80 - 251
TMhelix
252 - 274
outside
275 - 301
TMhelix
302 - 324
inside
325 - 330
TMhelix
331 - 350
outside
351 - 364
TMhelix
365 - 382
inside
383 - 385
Population Genetic Test Statistics
Pi
208.967335
Theta
171.082969
Tajima's D
-1.966376
CLR
0.420277
CSRT
0.0180490975451227
Interpretation
Possibly Positive selection
