Gene
KWMTBOMO15778
Pre Gene Modal
BGIBMGA002231
Annotation
PREDICTED:_F-box/LRR-repeat_protein_2_isoform_X1_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.043 Nuclear Reliability : 1.016 PlasmaMembrane Reliability : 1.526
Sequence
CDS
ATGGATATTTCCACTGATGTGTGGGGCCAGCTGGCTTTAGAGGCCTCGCAAGTGTACCTGACCGAGGGAGGCCAAGTGCGGAGTCCATTCGCGAGCACGACCATCGAAAAACTTCCGGACAAGGTTTTATTGAACATTTTCTCGTATCTAACACACCGTGAGATATGTCGTATGGCAACTGTTTGCAGACGATGGAGGATGGTTGCGTACGACACGAAATTGTGGACGCACGTTAGTTTGAGGCCGGAAATATCAGGATTGCACGTTAGTTCTCTGGAATCACTGTTAGCGCTGATATCAATAAGATTTGGACCGTCGCTGCGTTATATAGAGCTTCCAATAGAGCTGATAACCCACACAGTCCTCCACGAGTTGGCAGCTAAATGTCCTAATTTGACACACATGCTTCTTGATTTTAGTACTGCGATGCAACTCCACGACTTTTCGGAAATGCAAGCCTTCCCGACGAAGCTCCGCTACCTGTGCATCTGTCTGTCCGAGGTCATCTTCATGGAGGGATTCATGCGAAAGATTTACAATTTCATAAACGGCCTAGAGATATTGCATCTAATAGGCACGTACGAGAAGGTCGAAGAGGAAGAGGAAGAGATATACGAAGTGATAAACGTTCACAAATTGAAATCGGCGACTCCCAACCTAAGAGTCATCAATTTGTACGGGATAAACTTTGTCGACGACTCGCATATCGACGCGTTCAGTTCGAACTGTATACAGTTGGAATGTTTGGCGGTGAACTTTTGCAACAAGGTTACGGGATCCACCATGAAGACCTTGTTCCAGCGCTCGCGAAGACTCACCTGTCTCCTGATGAACGGATGCAGTCTTCGATCCGACTACGTAATGCAAGTGGAGTGGGAGAAATCCTCCGTTCAGGAATTGGATTTGACCGCCACAGACTTGTCGACCGAGTGCCTGATAGACATGCTCACCCGGATACCGAACCTGCGCTTTCTGAGCGCCGGCCAAATCAACGGGTTCAATGACTCTGTACTTAAAGCTTGGGTGGAAGCTGGGACTGCTAGGAACCTGGTGGCGCTGGACGTGGACTCGTCGGACAACCTGTCGGACGAAGCGCTGCACCGCTTCCTGTCGCGGCACGGCTCGCAGCTGCACGGGCTGGTGCTGGGCGGGATGCCGCACATCACCGACCAGCTGTGGCAGAGCGTGCTGCAGCTGCTCAACAACGCTAAGATCCTTATAATGGGAACTCAGGAGCGTCTCGGGATCAACATTCACGTTGACCAGCTGATGGATGGCATTGCGAACAGCTGCCCTAACTTGGAGCGCCTGGAGCTGCGCTGGGACCCGGAGAACCTGCGCTTCAGCGACAAGAGCCAGAAGGCGATCGACATACTCCGCGTCAAGTGTTTGAAACTAAAATGTTTAGTGCTGAGCGACGGCCGGTACTACGAAATCGTGAAGGCTAACTTCGAGCGCGCCGATCGCACTACCGTGGTCAGAACTTCCACAAATTGCCGAGTCTCAAACTACTATCTACTTTCAAATTACAACGATCTCATATTCAATTAA
Protein
MDISTDVWGQLALEASQVYLTEGGQVRSPFASTTIEKLPDKVLLNIFSYLTHREICRMATVCRRWRMVAYDTKLWTHVSLRPEISGLHVSSLESLLALISIRFGPSLRYIELPIELITHTVLHELAAKCPNLTHMLLDFSTAMQLHDFSEMQAFPTKLRYLCICLSEVIFMEGFMRKIYNFINGLEILHLIGTYEKVEEEEEEIYEVINVHKLKSATPNLRVINLYGINFVDDSHIDAFSSNCIQLECLAVNFCNKVTGSTMKTLFQRSRRLTCLLMNGCSLRSDYVMQVEWEKSSVQELDLTATDLSTECLIDMLTRIPNLRFLSAGQINGFNDSVLKAWVEAGTARNLVALDVDSSDNLSDEALHRFLSRHGSQLHGLVLGGMPHITDQLWQSVLQLLNNAKILIMGTQERLGINIHVDQLMDGIANSCPNLERLELRWDPENLRFSDKSQKAIDILRVKCLKLKCLVLSDGRYYEIVKANFERADRTTVVRTSTNCRVSNYYLLSNYNDLIFN
Summary
Uniprot
A0A2H1WQZ6
A0A2A4JDP1
A0A212EPC4
A0A2W1BFN9
H9IY98
E2CA99
+ More
A0A3S2NJS1 A0A0L7RBZ1 A0A088A816 A0A194QCF4 E2AVV5 A0A195FWK0 A0A1B6C1M8 A0A158P0Q2 A0A151WVY3 A0A195B1C0 A0A0C9R6S8 A0A0A9WKL7 A0A1B6IFQ1 A0A0A9WVT6 A0A023EX09 A0A1B6EYZ2 A0A1I8MTL9 A0A1B6LV92 A0A0K8UWI7 A0A1I8P024 W8BKK1 N6TY79 T1I131 A0A034VK39 A0A0Q9WT44 A0A0P9A327 Q9VAK7 Q8IGU5 B3P5H0 A0A0R1E918 A0A1W4UGZ1 A0A3B0KB20 I5AMY9 A0A158P0Q1 A0A0Q9W0G2 A0A3L8DCD5 A0A026VUZ1 A0A1J1J2U6 A0A1B6E1F8 A0A0C9PMU1 A0A151JAX4 J9K3Y1 A0A1B6HH84 A0A1B6D3M2 A0A2S2QHB7 A0A1B6HVJ8 A0A0C9QG95 A0A1B6H4G6 A0A1B6HL94 A0A2J7PUE0 A0A1W4WGB8 A0A224XP34 A0A182K0V3 A0A182UWF4 A0A182TNV3 A0A182WVJ4 A0A182KXT5 F5HLD8 A0A139WBR6 Q16RX7 B0WFD7 A0A182HE74 A0A182RBA0 A0A182NCH1 W8BW65 A0A0K8W989 A0A0A1XJW4 A0A1Y1N7E9 A0A1I8MTM0 A0A182YG40 T1PH57 A0A182F9K8 A0A1A9XTG5 A0A1B0BMI1 A0A182MC83 A0A067RRI1 W5JEQ4 A0A084VEN5 A0A1I8P004 A0A034VLQ8 A0A1I8NZZ7 A0A1J1J5X4 A0A0Q9WZ71 Q8T0K1 A0A2A3ENM2 B3MTE6 B4NBH6 A0A1A9WPN2 B4R044 B4HZA2 B4PQ29 A0A1W4U616 E9IYJ5 A0A182I973
A0A3S2NJS1 A0A0L7RBZ1 A0A088A816 A0A194QCF4 E2AVV5 A0A195FWK0 A0A1B6C1M8 A0A158P0Q2 A0A151WVY3 A0A195B1C0 A0A0C9R6S8 A0A0A9WKL7 A0A1B6IFQ1 A0A0A9WVT6 A0A023EX09 A0A1B6EYZ2 A0A1I8MTL9 A0A1B6LV92 A0A0K8UWI7 A0A1I8P024 W8BKK1 N6TY79 T1I131 A0A034VK39 A0A0Q9WT44 A0A0P9A327 Q9VAK7 Q8IGU5 B3P5H0 A0A0R1E918 A0A1W4UGZ1 A0A3B0KB20 I5AMY9 A0A158P0Q1 A0A0Q9W0G2 A0A3L8DCD5 A0A026VUZ1 A0A1J1J2U6 A0A1B6E1F8 A0A0C9PMU1 A0A151JAX4 J9K3Y1 A0A1B6HH84 A0A1B6D3M2 A0A2S2QHB7 A0A1B6HVJ8 A0A0C9QG95 A0A1B6H4G6 A0A1B6HL94 A0A2J7PUE0 A0A1W4WGB8 A0A224XP34 A0A182K0V3 A0A182UWF4 A0A182TNV3 A0A182WVJ4 A0A182KXT5 F5HLD8 A0A139WBR6 Q16RX7 B0WFD7 A0A182HE74 A0A182RBA0 A0A182NCH1 W8BW65 A0A0K8W989 A0A0A1XJW4 A0A1Y1N7E9 A0A1I8MTM0 A0A182YG40 T1PH57 A0A182F9K8 A0A1A9XTG5 A0A1B0BMI1 A0A182MC83 A0A067RRI1 W5JEQ4 A0A084VEN5 A0A1I8P004 A0A034VLQ8 A0A1I8NZZ7 A0A1J1J5X4 A0A0Q9WZ71 Q8T0K1 A0A2A3ENM2 B3MTE6 B4NBH6 A0A1A9WPN2 B4R044 B4HZA2 B4PQ29 A0A1W4U616 E9IYJ5 A0A182I973
Pubmed
22118469
28756777
19121390
20798317
26354079
21347285
+ More
25401762 26823975 25474469 25315136 24495485 23537049 25348373 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 15632085 30249741 24508170 20966253 12364791 14747013 17210077 18362917 19820115 17510324 26483478 25830018 28004739 25244985 24845553 20920257 23761445 24438588 26109357 26109356 21282665
25401762 26823975 25474469 25315136 24495485 23537049 25348373 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 15632085 30249741 24508170 20966253 12364791 14747013 17210077 18362917 19820115 17510324 26483478 25830018 28004739 25244985 24845553 20920257 23761445 24438588 26109357 26109356 21282665
EMBL
ODYU01010376
SOQ55448.1
NWSH01002164
NWSH01001833
PCG69010.1
PCG69959.1
+ More
AGBW02013496 OWR43326.1 KZ150129 PZC73081.1 BABH01020894 BABH01020895 BABH01020896 BABH01020897 GL453952 EFN75148.1 RSAL01000019 RVE52759.1 KQ414617 KOC68281.1 KQ459193 KPJ03147.1 GL443213 EFN62377.1 KQ981200 KYN45045.1 GEDC01029897 JAS07401.1 ADTU01005714 ADTU01005715 KQ982691 KYQ52030.1 KQ976667 KYM78261.1 GBYB01002466 JAG72233.1 GBHO01034617 GBHO01013411 GBRD01016125 GDHC01015916 JAG08987.1 JAG30193.1 JAG49701.1 JAQ02713.1 GECU01021955 GECU01001683 JAS85751.1 JAT06024.1 GBHO01034629 GBHO01013409 GBRD01016126 GDHC01003350 JAG08975.1 JAG30195.1 JAG49700.1 JAQ15279.1 GBBI01005025 JAC13687.1 GECZ01026503 GECZ01015390 JAS43266.1 JAS54379.1 GEBQ01012412 JAT27565.1 GDHF01021270 JAI31044.1 GAMC01009042 GAMC01009038 JAB97513.1 APGK01050349 KB741169 KB632203 ENN73336.1 ERL90065.1 ACPB03016231 GAKP01016465 JAC42487.1 CH964232 KRF99397.1 CH902623 KPU72917.1 AE014297 AAF56899.2 BT001595 AAN71350.1 CH954182 EDV53220.1 CM000160 KRK04193.1 OUUW01000007 SPP82866.1 CM000070 EIM52324.1 CH940652 KRF78618.1 QOIP01000010 RLU17991.1 KK108837 EZA46619.1 CVRI01000066 CRL06290.1 GEDC01005543 JAS31755.1 GBYB01002463 JAG72230.1 KQ979203 KYN22266.1 ABLF02039910 GECU01036927 GECU01033660 JAS70779.1 JAS74046.1 GEDC01028345 GEDC01017006 JAS08953.1 JAS20292.1 GGMS01007925 MBY77128.1 GECU01037453 GECU01028979 JAS70253.1 JAS78727.1 GBYB01002464 JAG72231.1 GECZ01000275 JAS69494.1 GECU01032351 JAS75355.1 NEVH01021199 PNF19950.1 GFTR01006635 JAW09791.1 AAAB01008888 EGK97099.1 KQ971372 KYB25377.1 CH477693 EAT37198.1 DS231916 EDS26154.1 JXUM01131183 KQ567678 KXJ69328.1 GAMC01009046 GAMC01009044 GAMC01009040 GAMC01009036 GAMC01009034 JAB97511.1 GDHF01012170 GDHF01004705 JAI40144.1 JAI47609.1 GBXI01003112 JAD11180.1 GEZM01012970 JAV92840.1 KA647223 AFP61852.1 JXJN01016937 JXJN01016938 AXCM01000042 KK852509 KDR22374.1 ADMH02001608 ETN61823.1 ATLV01012285 KE524778 KFB36429.1 GAKP01016466 GAKP01016464 JAC42488.1 CRL06289.1 CH933806 KRG01211.1 AY069210 AAL39355.1 AAN14173.1 KZ288203 PBC33365.1 EDV30536.1 EDW81140.1 CM000364 EDX14850.1 CH480819 EDW53359.1 EDW98298.1 KRK04194.1 KRK04195.1 GL767018 EFZ14370.1 APCN01003212
AGBW02013496 OWR43326.1 KZ150129 PZC73081.1 BABH01020894 BABH01020895 BABH01020896 BABH01020897 GL453952 EFN75148.1 RSAL01000019 RVE52759.1 KQ414617 KOC68281.1 KQ459193 KPJ03147.1 GL443213 EFN62377.1 KQ981200 KYN45045.1 GEDC01029897 JAS07401.1 ADTU01005714 ADTU01005715 KQ982691 KYQ52030.1 KQ976667 KYM78261.1 GBYB01002466 JAG72233.1 GBHO01034617 GBHO01013411 GBRD01016125 GDHC01015916 JAG08987.1 JAG30193.1 JAG49701.1 JAQ02713.1 GECU01021955 GECU01001683 JAS85751.1 JAT06024.1 GBHO01034629 GBHO01013409 GBRD01016126 GDHC01003350 JAG08975.1 JAG30195.1 JAG49700.1 JAQ15279.1 GBBI01005025 JAC13687.1 GECZ01026503 GECZ01015390 JAS43266.1 JAS54379.1 GEBQ01012412 JAT27565.1 GDHF01021270 JAI31044.1 GAMC01009042 GAMC01009038 JAB97513.1 APGK01050349 KB741169 KB632203 ENN73336.1 ERL90065.1 ACPB03016231 GAKP01016465 JAC42487.1 CH964232 KRF99397.1 CH902623 KPU72917.1 AE014297 AAF56899.2 BT001595 AAN71350.1 CH954182 EDV53220.1 CM000160 KRK04193.1 OUUW01000007 SPP82866.1 CM000070 EIM52324.1 CH940652 KRF78618.1 QOIP01000010 RLU17991.1 KK108837 EZA46619.1 CVRI01000066 CRL06290.1 GEDC01005543 JAS31755.1 GBYB01002463 JAG72230.1 KQ979203 KYN22266.1 ABLF02039910 GECU01036927 GECU01033660 JAS70779.1 JAS74046.1 GEDC01028345 GEDC01017006 JAS08953.1 JAS20292.1 GGMS01007925 MBY77128.1 GECU01037453 GECU01028979 JAS70253.1 JAS78727.1 GBYB01002464 JAG72231.1 GECZ01000275 JAS69494.1 GECU01032351 JAS75355.1 NEVH01021199 PNF19950.1 GFTR01006635 JAW09791.1 AAAB01008888 EGK97099.1 KQ971372 KYB25377.1 CH477693 EAT37198.1 DS231916 EDS26154.1 JXUM01131183 KQ567678 KXJ69328.1 GAMC01009046 GAMC01009044 GAMC01009040 GAMC01009036 GAMC01009034 JAB97511.1 GDHF01012170 GDHF01004705 JAI40144.1 JAI47609.1 GBXI01003112 JAD11180.1 GEZM01012970 JAV92840.1 KA647223 AFP61852.1 JXJN01016937 JXJN01016938 AXCM01000042 KK852509 KDR22374.1 ADMH02001608 ETN61823.1 ATLV01012285 KE524778 KFB36429.1 GAKP01016466 GAKP01016464 JAC42488.1 CRL06289.1 CH933806 KRG01211.1 AY069210 AAL39355.1 AAN14173.1 KZ288203 PBC33365.1 EDV30536.1 EDW81140.1 CM000364 EDX14850.1 CH480819 EDW53359.1 EDW98298.1 KRK04194.1 KRK04195.1 GL767018 EFZ14370.1 APCN01003212
Proteomes
UP000218220
UP000007151
UP000005204
UP000008237
UP000283053
UP000053825
+ More
UP000005203 UP000053268 UP000000311 UP000078541 UP000005205 UP000075809 UP000078540 UP000095301 UP000095300 UP000019118 UP000030742 UP000015103 UP000007798 UP000007801 UP000000803 UP000008711 UP000002282 UP000192221 UP000268350 UP000001819 UP000008792 UP000279307 UP000053097 UP000183832 UP000078492 UP000007819 UP000235965 UP000192223 UP000075881 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000007266 UP000008820 UP000002320 UP000069940 UP000249989 UP000075900 UP000075884 UP000076408 UP000069272 UP000092443 UP000092460 UP000075883 UP000027135 UP000000673 UP000030765 UP000009192 UP000242457 UP000091820 UP000000304 UP000001292 UP000075840
UP000005203 UP000053268 UP000000311 UP000078541 UP000005205 UP000075809 UP000078540 UP000095301 UP000095300 UP000019118 UP000030742 UP000015103 UP000007798 UP000007801 UP000000803 UP000008711 UP000002282 UP000192221 UP000268350 UP000001819 UP000008792 UP000279307 UP000053097 UP000183832 UP000078492 UP000007819 UP000235965 UP000192223 UP000075881 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000007266 UP000008820 UP000002320 UP000069940 UP000249989 UP000075900 UP000075884 UP000076408 UP000069272 UP000092443 UP000092460 UP000075883 UP000027135 UP000000673 UP000030765 UP000009192 UP000242457 UP000091820 UP000000304 UP000001292 UP000075840
Interpro
Gene 3D
ProteinModelPortal
A0A2H1WQZ6
A0A2A4JDP1
A0A212EPC4
A0A2W1BFN9
H9IY98
E2CA99
+ More
A0A3S2NJS1 A0A0L7RBZ1 A0A088A816 A0A194QCF4 E2AVV5 A0A195FWK0 A0A1B6C1M8 A0A158P0Q2 A0A151WVY3 A0A195B1C0 A0A0C9R6S8 A0A0A9WKL7 A0A1B6IFQ1 A0A0A9WVT6 A0A023EX09 A0A1B6EYZ2 A0A1I8MTL9 A0A1B6LV92 A0A0K8UWI7 A0A1I8P024 W8BKK1 N6TY79 T1I131 A0A034VK39 A0A0Q9WT44 A0A0P9A327 Q9VAK7 Q8IGU5 B3P5H0 A0A0R1E918 A0A1W4UGZ1 A0A3B0KB20 I5AMY9 A0A158P0Q1 A0A0Q9W0G2 A0A3L8DCD5 A0A026VUZ1 A0A1J1J2U6 A0A1B6E1F8 A0A0C9PMU1 A0A151JAX4 J9K3Y1 A0A1B6HH84 A0A1B6D3M2 A0A2S2QHB7 A0A1B6HVJ8 A0A0C9QG95 A0A1B6H4G6 A0A1B6HL94 A0A2J7PUE0 A0A1W4WGB8 A0A224XP34 A0A182K0V3 A0A182UWF4 A0A182TNV3 A0A182WVJ4 A0A182KXT5 F5HLD8 A0A139WBR6 Q16RX7 B0WFD7 A0A182HE74 A0A182RBA0 A0A182NCH1 W8BW65 A0A0K8W989 A0A0A1XJW4 A0A1Y1N7E9 A0A1I8MTM0 A0A182YG40 T1PH57 A0A182F9K8 A0A1A9XTG5 A0A1B0BMI1 A0A182MC83 A0A067RRI1 W5JEQ4 A0A084VEN5 A0A1I8P004 A0A034VLQ8 A0A1I8NZZ7 A0A1J1J5X4 A0A0Q9WZ71 Q8T0K1 A0A2A3ENM2 B3MTE6 B4NBH6 A0A1A9WPN2 B4R044 B4HZA2 B4PQ29 A0A1W4U616 E9IYJ5 A0A182I973
A0A3S2NJS1 A0A0L7RBZ1 A0A088A816 A0A194QCF4 E2AVV5 A0A195FWK0 A0A1B6C1M8 A0A158P0Q2 A0A151WVY3 A0A195B1C0 A0A0C9R6S8 A0A0A9WKL7 A0A1B6IFQ1 A0A0A9WVT6 A0A023EX09 A0A1B6EYZ2 A0A1I8MTL9 A0A1B6LV92 A0A0K8UWI7 A0A1I8P024 W8BKK1 N6TY79 T1I131 A0A034VK39 A0A0Q9WT44 A0A0P9A327 Q9VAK7 Q8IGU5 B3P5H0 A0A0R1E918 A0A1W4UGZ1 A0A3B0KB20 I5AMY9 A0A158P0Q1 A0A0Q9W0G2 A0A3L8DCD5 A0A026VUZ1 A0A1J1J2U6 A0A1B6E1F8 A0A0C9PMU1 A0A151JAX4 J9K3Y1 A0A1B6HH84 A0A1B6D3M2 A0A2S2QHB7 A0A1B6HVJ8 A0A0C9QG95 A0A1B6H4G6 A0A1B6HL94 A0A2J7PUE0 A0A1W4WGB8 A0A224XP34 A0A182K0V3 A0A182UWF4 A0A182TNV3 A0A182WVJ4 A0A182KXT5 F5HLD8 A0A139WBR6 Q16RX7 B0WFD7 A0A182HE74 A0A182RBA0 A0A182NCH1 W8BW65 A0A0K8W989 A0A0A1XJW4 A0A1Y1N7E9 A0A1I8MTM0 A0A182YG40 T1PH57 A0A182F9K8 A0A1A9XTG5 A0A1B0BMI1 A0A182MC83 A0A067RRI1 W5JEQ4 A0A084VEN5 A0A1I8P004 A0A034VLQ8 A0A1I8NZZ7 A0A1J1J5X4 A0A0Q9WZ71 Q8T0K1 A0A2A3ENM2 B3MTE6 B4NBH6 A0A1A9WPN2 B4R044 B4HZA2 B4PQ29 A0A1W4U616 E9IYJ5 A0A182I973
PDB
2AST
E-value=2.04922e-05,
Score=116
Ontologies
GO
PANTHER
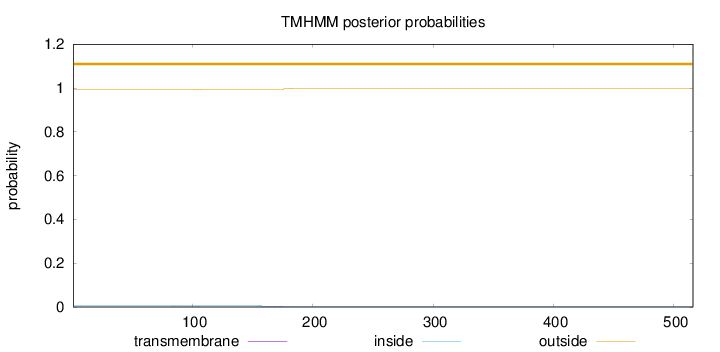
Topology
Length:
516
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10297
Exp number, first 60 AAs:
0.00147
Total prob of N-in:
0.00767
outside
1 - 516
Population Genetic Test Statistics
Pi
270.487925
Theta
174.319436
Tajima's D
1.197615
CLR
0.420277
CSRT
0.715914204289785
Interpretation
Uncertain
