Gene
KWMTBOMO15776
Pre Gene Modal
BGIBMGA002234
Annotation
PREDICTED:_protein_glass_[Papilio_xuthus]
Full name
Protein glass
Transcription factor
Location in the cell
Nuclear Reliability : 4.612
Sequence
CDS
ATGCAAGAAGGACATAAAGACCAGGGTGCGGGAGGCGGGTTCATAATGAACCACCAAACATCAATGCACCCACAGGACTACGATTATTCTGCTTGTGTGAGCGCGGGGAAAGTTCAGGTGGGAGCTTTAAGCGGATGTTCGGAAGACGACGAACAAAAGCCTAACTTATGTCGTATTTGCGGTAAGACTTACGCCCGGCCTAGCACTTTGAAGACCCACCTTCGGACGCATTCGGGTGAGCGGCCGTATCGCTGTGGGGATTGCAACAAGAGCTTCTCCCAGGCTGCTAACTTGACGGCTCATGTCAGGACGCATACGGGACAGAAGCCTTTCAGATGTCCTATCTGCGATCGAAGGTTTAGCCAAAGCTCCAGCGTGACGACACACATGCGGACGCATTCAGGGGAGCGACCCTATCAATGCAGATCATGCAAAAAAGCATTTTCAGATAGCTCGACGTTGACCAAGCACCTCCGGATACACTCCGGAGAGAAACCTTATCAATGCAAACTCTGTTTATTGAGATTTTCGCAATCTGGCAATCTGAACCGTCACATGCGAGTCCACGGTAACATGTCGGCAGGGATGCTCGGCTGA
Protein
MQEGHKDQGAGGGFIMNHQTSMHPQDYDYSACVSAGKVQVGALSGCSEDDEQKPNLCRICGKTYARPSTLKTHLRTHSGERPYRCGDCNKSFSQAANLTAHVRTHTGQKPFRCPICDRRFSQSSSVTTHMRTHSGERPYQCRSCKKAFSDSSTLTKHLRIHSGEKPYQCKLCLLRFSQSGNLNRHMRVHGNMSAGMLG
Summary
Description
Transcription factor required for gene expression specific to photoreceptor cells.
Keywords
Alternative splicing
Complete proteome
DNA-binding
Metal-binding
Nucleus
Reference proteome
Repeat
Sensory transduction
Transcription
Transcription regulation
Vision
Zinc
Zinc-finger
Feature
chain Protein glass
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A2H1WGB0
A0A194QCK3
A0A0N1INI8
A0A212EPA9
A0A182R8E9
A0A1W4XKU7
+ More
A0A0A1WJX5 A0A182SV13 W8BC86 D6X386 Q7Z1F3 A0A1W4XJY2 A0A182J6D3 A0A0P4WLG9 A0A182LZI8 A0A182QEC2 A0A1B0BJC9 A0A182HVH0 A0A182WUX9 A0A1S4GKN6 A0A1S3DFF5 A0A240PLZ4 E0VZC5 Q7PQ71 A0A1I8PZC2 A0A1A9XZA7 N6UB25 A0A1A9ZXM5 A0A1B0F9Q2 A0A034V7D5 W8B0H6 A0A0K8U0A7 A0A1A9VEQ7 A0A1I8N8P7 A0A1A9W3I8 B0WGR9 A0A0C9RJI0 P13360 B3NZ66 B4I295 A0A1W4U8E4 B4QUF3 A0A0L0C951 A0A0M5J936 Q24732 A0A0B4KGX3 B4G5D2 A0A195D0J0 F4X7S6 A0A3L8DSB0 A0A3B0K1A5 A0A1J1J1G1 A0A3S5CZE0 B4PLB6 H9IYA1 B4K932 B4N9D5 A0A2S2PFP8 A0A195E624 A0A158NPF2 B3LWA4 A0A195B9A9 A0A151WRJ7 K7INJ2 A0A195ETV9 A0A2H8TJR6 J9K6U8 Q299L5 A0A0L8GP86 T1IIQ5 R7TLC5 A0A2A3E3M7 A0A087ZWL4 A0A226DFR0 A0A154PJV1 A0A310S8X9 V4B2M4 A0A0P6HKI8 A0A0P6CM08 K1QUI9 A0A164VPL5 A0A0P5W7U5 A0A336LRC9 A0A0L7R2H0 A0A2T7PAV1 D2XMT2 A0A1B0GH94 A0A3S1B573 B7QJK4 V9PPG3
A0A0A1WJX5 A0A182SV13 W8BC86 D6X386 Q7Z1F3 A0A1W4XJY2 A0A182J6D3 A0A0P4WLG9 A0A182LZI8 A0A182QEC2 A0A1B0BJC9 A0A182HVH0 A0A182WUX9 A0A1S4GKN6 A0A1S3DFF5 A0A240PLZ4 E0VZC5 Q7PQ71 A0A1I8PZC2 A0A1A9XZA7 N6UB25 A0A1A9ZXM5 A0A1B0F9Q2 A0A034V7D5 W8B0H6 A0A0K8U0A7 A0A1A9VEQ7 A0A1I8N8P7 A0A1A9W3I8 B0WGR9 A0A0C9RJI0 P13360 B3NZ66 B4I295 A0A1W4U8E4 B4QUF3 A0A0L0C951 A0A0M5J936 Q24732 A0A0B4KGX3 B4G5D2 A0A195D0J0 F4X7S6 A0A3L8DSB0 A0A3B0K1A5 A0A1J1J1G1 A0A3S5CZE0 B4PLB6 H9IYA1 B4K932 B4N9D5 A0A2S2PFP8 A0A195E624 A0A158NPF2 B3LWA4 A0A195B9A9 A0A151WRJ7 K7INJ2 A0A195ETV9 A0A2H8TJR6 J9K6U8 Q299L5 A0A0L8GP86 T1IIQ5 R7TLC5 A0A2A3E3M7 A0A087ZWL4 A0A226DFR0 A0A154PJV1 A0A310S8X9 V4B2M4 A0A0P6HKI8 A0A0P6CM08 K1QUI9 A0A164VPL5 A0A0P5W7U5 A0A336LRC9 A0A0L7R2H0 A0A2T7PAV1 D2XMT2 A0A1B0GH94 A0A3S1B573 B7QJK4 V9PPG3
Pubmed
26354079
22118469
25830018
24495485
18362917
19820115
+ More
15081356 12364791 20566863 23537049 25348373 25315136 2770860 10731132 12537572 12537569 7604032 17994087 26108605 12537568 12537573 12537574 16110336 17569856 17569867 21719571 30249741 17550304 19121390 21347285 20075255 15632085 23254933 22992520 24337300
15081356 12364791 20566863 23537049 25348373 25315136 2770860 10731132 12537572 12537569 7604032 17994087 26108605 12537568 12537573 12537574 16110336 17569856 17569867 21719571 30249741 17550304 19121390 21347285 20075255 15632085 23254933 22992520 24337300
EMBL
ODYU01008467
SOQ52101.1
KQ459193
KPJ03149.1
KQ461188
KPJ07327.1
+ More
AGBW02013496 OWR43328.1 GBXI01015301 JAC98990.1 GAMC01011947 JAB94608.1 KQ971372 EFA10761.1 AY293621 AAP46162.1 GDRN01037995 JAI67273.1 AXCM01000299 AXCM01000300 AXCN02001183 JXJN01015358 JXJN01015359 APCN01005776 APCN01005777 AAAB01008898 DS235851 EEB18731.1 EAA09224.6 APGK01041747 APGK01041748 KB740997 ENN75837.1 CCAG010019856 GAKP01020910 JAC38042.1 GAMC01011950 JAB94605.1 GDHF01032162 JAI20152.1 DS231928 EDS27145.1 GBYB01008360 JAG78127.1 X15400 AE014297 AY060322 CH954181 EDV48608.1 CH480820 EDW54652.1 CM000364 EDX12453.1 JRES01000835 KNC27919.1 CP012526 ALC46098.1 U39746 CH940652 AGB96088.1 CH479179 EDW24798.1 KQ977004 KYN06433.1 GL888890 EGI57499.1 QOIP01000005 RLU22648.1 OUUW01000013 SPP88014.1 CVRI01000066 CRL06176.1 LR129951 VDH80593.1 CM000160 EDW95898.2 BABH01020889 CH933806 EDW14445.2 CH964232 EDW80568.2 GGMR01015641 MBY28260.1 KQ979608 KYN20304.1 ADTU01022477 ADTU01022478 CH902617 EDV42682.2 KQ976542 KYM81121.1 KQ982807 KYQ50427.1 KQ981965 KYN31678.1 GFXV01002465 MBW14270.1 ABLF02024975 ABLF02024982 ABLF02026914 ABLF02026922 ABLF02046262 ABLF02049869 CM000070 EAL27688.3 KQ420924 KOF78841.1 JH430212 AMQN01012256 KB309404 ELT94479.1 KZ288438 PBC25749.1 LNIX01000020 OXA44043.1 KQ434938 KZC12161.1 KQ768431 OAD53168.1 KB203854 ESO82734.1 GDIQ01017738 JAN76999.1 GDIP01000177 JAN03539.1 JH815769 EKC25221.1 LRGB01001361 KZS12527.1 GDIP01103332 JAM00383.1 UFQT01000116 SSX20345.1 KQ414666 KOC65072.1 PZQS01000005 PVD30554.1 GU211202 ADB22419.1 AJWK01004101 AJWK01004102 RQTK01000406 RUS80191.1 ABJB010072605 ABJB010191938 ABJB010215013 ABJB010372482 ABJB010542432 ABJB010587243 ABJB010953027 ABJB011031429 DS953043 EEC19026.1 KF317326 AHA51260.1
AGBW02013496 OWR43328.1 GBXI01015301 JAC98990.1 GAMC01011947 JAB94608.1 KQ971372 EFA10761.1 AY293621 AAP46162.1 GDRN01037995 JAI67273.1 AXCM01000299 AXCM01000300 AXCN02001183 JXJN01015358 JXJN01015359 APCN01005776 APCN01005777 AAAB01008898 DS235851 EEB18731.1 EAA09224.6 APGK01041747 APGK01041748 KB740997 ENN75837.1 CCAG010019856 GAKP01020910 JAC38042.1 GAMC01011950 JAB94605.1 GDHF01032162 JAI20152.1 DS231928 EDS27145.1 GBYB01008360 JAG78127.1 X15400 AE014297 AY060322 CH954181 EDV48608.1 CH480820 EDW54652.1 CM000364 EDX12453.1 JRES01000835 KNC27919.1 CP012526 ALC46098.1 U39746 CH940652 AGB96088.1 CH479179 EDW24798.1 KQ977004 KYN06433.1 GL888890 EGI57499.1 QOIP01000005 RLU22648.1 OUUW01000013 SPP88014.1 CVRI01000066 CRL06176.1 LR129951 VDH80593.1 CM000160 EDW95898.2 BABH01020889 CH933806 EDW14445.2 CH964232 EDW80568.2 GGMR01015641 MBY28260.1 KQ979608 KYN20304.1 ADTU01022477 ADTU01022478 CH902617 EDV42682.2 KQ976542 KYM81121.1 KQ982807 KYQ50427.1 KQ981965 KYN31678.1 GFXV01002465 MBW14270.1 ABLF02024975 ABLF02024982 ABLF02026914 ABLF02026922 ABLF02046262 ABLF02049869 CM000070 EAL27688.3 KQ420924 KOF78841.1 JH430212 AMQN01012256 KB309404 ELT94479.1 KZ288438 PBC25749.1 LNIX01000020 OXA44043.1 KQ434938 KZC12161.1 KQ768431 OAD53168.1 KB203854 ESO82734.1 GDIQ01017738 JAN76999.1 GDIP01000177 JAN03539.1 JH815769 EKC25221.1 LRGB01001361 KZS12527.1 GDIP01103332 JAM00383.1 UFQT01000116 SSX20345.1 KQ414666 KOC65072.1 PZQS01000005 PVD30554.1 GU211202 ADB22419.1 AJWK01004101 AJWK01004102 RQTK01000406 RUS80191.1 ABJB010072605 ABJB010191938 ABJB010215013 ABJB010372482 ABJB010542432 ABJB010587243 ABJB010953027 ABJB011031429 DS953043 EEC19026.1 KF317326 AHA51260.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000075900
UP000192223
UP000075901
+ More
UP000007266 UP000075880 UP000075883 UP000075886 UP000092460 UP000075840 UP000076407 UP000079169 UP000075885 UP000009046 UP000007062 UP000095300 UP000092443 UP000019118 UP000092445 UP000092444 UP000078200 UP000095301 UP000091820 UP000002320 UP000000803 UP000008711 UP000001292 UP000192221 UP000000304 UP000037069 UP000092553 UP000008792 UP000008744 UP000078542 UP000007755 UP000279307 UP000268350 UP000183832 UP000002282 UP000005204 UP000009192 UP000007798 UP000078492 UP000005205 UP000007801 UP000078540 UP000075809 UP000002358 UP000078541 UP000007819 UP000001819 UP000053454 UP000014760 UP000242457 UP000005203 UP000198287 UP000076502 UP000030746 UP000005408 UP000076858 UP000053825 UP000245119 UP000092461 UP000271974 UP000001555
UP000007266 UP000075880 UP000075883 UP000075886 UP000092460 UP000075840 UP000076407 UP000079169 UP000075885 UP000009046 UP000007062 UP000095300 UP000092443 UP000019118 UP000092445 UP000092444 UP000078200 UP000095301 UP000091820 UP000002320 UP000000803 UP000008711 UP000001292 UP000192221 UP000000304 UP000037069 UP000092553 UP000008792 UP000008744 UP000078542 UP000007755 UP000279307 UP000268350 UP000183832 UP000002282 UP000005204 UP000009192 UP000007798 UP000078492 UP000005205 UP000007801 UP000078540 UP000075809 UP000002358 UP000078541 UP000007819 UP000001819 UP000053454 UP000014760 UP000242457 UP000005203 UP000198287 UP000076502 UP000030746 UP000005408 UP000076858 UP000053825 UP000245119 UP000092461 UP000271974 UP000001555
Interpro
ProteinModelPortal
A0A2H1WGB0
A0A194QCK3
A0A0N1INI8
A0A212EPA9
A0A182R8E9
A0A1W4XKU7
+ More
A0A0A1WJX5 A0A182SV13 W8BC86 D6X386 Q7Z1F3 A0A1W4XJY2 A0A182J6D3 A0A0P4WLG9 A0A182LZI8 A0A182QEC2 A0A1B0BJC9 A0A182HVH0 A0A182WUX9 A0A1S4GKN6 A0A1S3DFF5 A0A240PLZ4 E0VZC5 Q7PQ71 A0A1I8PZC2 A0A1A9XZA7 N6UB25 A0A1A9ZXM5 A0A1B0F9Q2 A0A034V7D5 W8B0H6 A0A0K8U0A7 A0A1A9VEQ7 A0A1I8N8P7 A0A1A9W3I8 B0WGR9 A0A0C9RJI0 P13360 B3NZ66 B4I295 A0A1W4U8E4 B4QUF3 A0A0L0C951 A0A0M5J936 Q24732 A0A0B4KGX3 B4G5D2 A0A195D0J0 F4X7S6 A0A3L8DSB0 A0A3B0K1A5 A0A1J1J1G1 A0A3S5CZE0 B4PLB6 H9IYA1 B4K932 B4N9D5 A0A2S2PFP8 A0A195E624 A0A158NPF2 B3LWA4 A0A195B9A9 A0A151WRJ7 K7INJ2 A0A195ETV9 A0A2H8TJR6 J9K6U8 Q299L5 A0A0L8GP86 T1IIQ5 R7TLC5 A0A2A3E3M7 A0A087ZWL4 A0A226DFR0 A0A154PJV1 A0A310S8X9 V4B2M4 A0A0P6HKI8 A0A0P6CM08 K1QUI9 A0A164VPL5 A0A0P5W7U5 A0A336LRC9 A0A0L7R2H0 A0A2T7PAV1 D2XMT2 A0A1B0GH94 A0A3S1B573 B7QJK4 V9PPG3
A0A0A1WJX5 A0A182SV13 W8BC86 D6X386 Q7Z1F3 A0A1W4XJY2 A0A182J6D3 A0A0P4WLG9 A0A182LZI8 A0A182QEC2 A0A1B0BJC9 A0A182HVH0 A0A182WUX9 A0A1S4GKN6 A0A1S3DFF5 A0A240PLZ4 E0VZC5 Q7PQ71 A0A1I8PZC2 A0A1A9XZA7 N6UB25 A0A1A9ZXM5 A0A1B0F9Q2 A0A034V7D5 W8B0H6 A0A0K8U0A7 A0A1A9VEQ7 A0A1I8N8P7 A0A1A9W3I8 B0WGR9 A0A0C9RJI0 P13360 B3NZ66 B4I295 A0A1W4U8E4 B4QUF3 A0A0L0C951 A0A0M5J936 Q24732 A0A0B4KGX3 B4G5D2 A0A195D0J0 F4X7S6 A0A3L8DSB0 A0A3B0K1A5 A0A1J1J1G1 A0A3S5CZE0 B4PLB6 H9IYA1 B4K932 B4N9D5 A0A2S2PFP8 A0A195E624 A0A158NPF2 B3LWA4 A0A195B9A9 A0A151WRJ7 K7INJ2 A0A195ETV9 A0A2H8TJR6 J9K6U8 Q299L5 A0A0L8GP86 T1IIQ5 R7TLC5 A0A2A3E3M7 A0A087ZWL4 A0A226DFR0 A0A154PJV1 A0A310S8X9 V4B2M4 A0A0P6HKI8 A0A0P6CM08 K1QUI9 A0A164VPL5 A0A0P5W7U5 A0A336LRC9 A0A0L7R2H0 A0A2T7PAV1 D2XMT2 A0A1B0GH94 A0A3S1B573 B7QJK4 V9PPG3
PDB
2I13
E-value=2.08855e-31,
Score=335
Ontologies
GO
GO:0003676
GO:0003677
GO:0006355
GO:0001751
GO:0060086
GO:0043153
GO:0046872
GO:0005634
GO:0010114
GO:0009649
GO:0007601
GO:0007605
GO:0046530
GO:0045944
GO:0035271
GO:0001752
GO:0001745
GO:0000981
GO:0050896
GO:0016021
GO:0005524
GO:0042626
GO:0006468
GO:0008270
GO:0005215
GO:0006270
GO:0042555
GO:0003707
GO:0006281
GO:0006259
PANTHER
Topology
Subcellular location
Nucleus
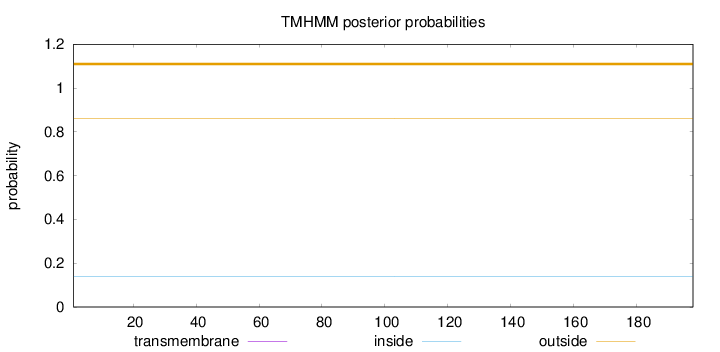
Length:
198
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.13784
outside
1 - 198
Population Genetic Test Statistics
Pi
22.436881
Theta
27.989591
Tajima's D
-0.585555
CLR
0.487542
CSRT
0.223738813059347
Interpretation
Uncertain
