Gene
KWMTBOMO15758 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002241
Annotation
vacuolar_ATP_synthase_subunit_B_[Bombyx_mori]
Full name
V-type proton ATPase subunit B
Alternative Name
Vacuolar proton pump subunit B
V-ATPase 55 kDa subunit
V-ATPase 55 kDa subunit
Location in the cell
Cytoplasmic Reliability : 1.997
Sequence
CDS
ATGGCAAAGGTGATCTCTCACGCCCAGGCGACCAAGGAACATGTCCTGGCAGTCTCCAGGGACTTCATTTCCCAGCCCAGGCTCACTTACAAGACTGTGTCTGGTGTAAACGGTCCTCTCGTCATCTTGGATGAAGTCAAGTTCCCCAAGTTCTCTGAGATCGTACAGCTAAAGCTTGCAGATGGAACCCTTCGTTCTGGTCAGGTACTCGAGGTCAGCGGCTCCAAAGCCGTCGTCCAAGTGTTCGAAGGGACATCAGGTATTGACGCGAAGAACACACTCTGCGAGTTCACGGGTGACATTCTTCGTACACCGGTCTCTGAAGACATGTTGGGTCGCGTGTTCAACGGTTCCGGCAAACCCATCGACAAGGGTCCCCCAATCTTGGCCGAGGACTTCTTGGACATCCAGGGTCAGCCCATCAACCCATGGTCACGTATCTACCCAGAGGAGATGATCCAAACTGGTATCTCCGCTATTGATGTGATGAACTCTATTGCCCGTGGTCAGAAGATCCCCATCTTCTCGGCCGCCGGTTTGCCTCACAATGAAATCGCCGCCCAGATCTGTAGACAGGCCGGTCTTGTCAAGGTCCCCGGCAAGTCAGTCTTGGACGACCACGAGGACAACTTCGCCATTGTGTTCGCCGCTATGGGTGTCAACATGGAAACGGCCAGATTCTTCAAGCAGGACTTTGAAGAGAACGGTTCCATGGAGAACGTGTGTCTCTTCTTGAACTTGGCCAACGATCCTACCATCGAGAGAATTATTACACCTCGTCTCGCCCTGACCGCCGCCGAGTTCTTGGCTTACCAGTGTGAGGTGAGTGTGTGTGAGAGAGAGAGACAATACACTCTATTGCACTCCAAAACACAATTTACATTCAAAAACAGTCAAAAAGGCGGTCTTATCGCTAAAAGCGATCTCTTCCACACCCCCTTTTAA
Protein
MAKVISHAQATKEHVLAVSRDFISQPRLTYKTVSGVNGPLVILDEVKFPKFSEIVQLKLADGTLRSGQVLEVSGSKAVVQVFEGTSGIDAKNTLCEFTGDILRTPVSEDMLGRVFNGSGKPIDKGPPILAEDFLDIQGQPINPWSRIYPEEMIQTGISAIDVMNSIARGQKIPIFSAAGLPHNEIAAQICRQAGLVKVPGKSVLDDHEDNFAIVFAAMGVNMETARFFKQDFEENGSMENVCLFLNLANDPTIERIITPRLALTAAEFLAYQCEVSVCERERQYTLLHSKTQFTFKNSQKGGLIAKSDLFHTPF
Summary
Subunit
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (main components: subunits A, B, C, D, E, and F) attached to an integral membrane V0 proton pore complex (main component: the proteolipid protein).
Similarity
Belongs to the ATPase alpha/beta chains family.
Keywords
Hydrogen ion transport
Hydrolase
Ion transport
Transport
Complete proteome
Reference proteome
Feature
chain V-type proton ATPase subunit B
Uniprot
A1E4A8
B3VBE3
A0A3S2NP74
A0A1S5XVQ5
A0A194QJG9
P31401
+ More
A0A0N1PFD7 S4PMQ2 I4DJA0 A0A212ERZ3 E7BZ73 A0A2A4IUP6 P31410 A0A0L0C0P8 A0A1I8P324 A0A1L8EEN3 W8BUW3 T1PF56 A0A0K8VVU7 A0A2Z4GWT6 A0A151WMF7 A0A0K8W0S0 A0A034VT14 A0A0A1X2S7 A0A195CSA0 A0A151IZY9 A0A2R7WZ85 A0A195BP22 A0A158NVJ8 A0A1B6KH42 E2AP90 A0A195FY38 A0A1B6HKP9 A0A154P289 A0A3B0K915 F4WLD7 A0A1J1HPL7 E2BNJ3 A0A026X5Q0 A0A3B0KFZ3 A0A1Q3FCM0 B4GN04 Q29BS3 O44427 B4MC74 A0A1W4UPV6 E1JIJ5 B4PPC2 A0A0M3QXN3 B3MST7 B4NHZ6 B3P492 A0A1S6M253 A0A0K8TQ02 B4K7G7 P31409 B4QS89 A0A0L7RJE6 A0A1A9YSX7 A0A1B0B2L4 B4HGV7 A0A1Y1L4S1 A0A1W4XVH5 A0A3G1SUY5 V5GPH2 A0A161MEN8 B4JGS2 A0A1A9VFV9 D6X1R1 A0A1B1UZQ0 D3TSC7 A0A232FGG3 A0A1S6M248 A0A0T6B7V7 A0A069DUZ5 A0A1L8DFJ9 A0A1L1YME0 A0A0N7Z9E6 A0A0V0G3F6 A0A2J7RFL5 T1I122 A0A1I9WLE3 A0A1B0CIV0 A0A023EUA8 A0A182K3I9 A0A182VYQ1 A0A084VXP8 A0A2A3EDR1 V9IKE7 A0A088ADH1 A0A182P806 A0A1A9ZZ34 Q9XYC8 A0A1D1YDC1 J3JVB5 E0VJX6 A0A0U1XFD8 M9TFX8 A0A182XHK3 A0A182MD68
A0A0N1PFD7 S4PMQ2 I4DJA0 A0A212ERZ3 E7BZ73 A0A2A4IUP6 P31410 A0A0L0C0P8 A0A1I8P324 A0A1L8EEN3 W8BUW3 T1PF56 A0A0K8VVU7 A0A2Z4GWT6 A0A151WMF7 A0A0K8W0S0 A0A034VT14 A0A0A1X2S7 A0A195CSA0 A0A151IZY9 A0A2R7WZ85 A0A195BP22 A0A158NVJ8 A0A1B6KH42 E2AP90 A0A195FY38 A0A1B6HKP9 A0A154P289 A0A3B0K915 F4WLD7 A0A1J1HPL7 E2BNJ3 A0A026X5Q0 A0A3B0KFZ3 A0A1Q3FCM0 B4GN04 Q29BS3 O44427 B4MC74 A0A1W4UPV6 E1JIJ5 B4PPC2 A0A0M3QXN3 B3MST7 B4NHZ6 B3P492 A0A1S6M253 A0A0K8TQ02 B4K7G7 P31409 B4QS89 A0A0L7RJE6 A0A1A9YSX7 A0A1B0B2L4 B4HGV7 A0A1Y1L4S1 A0A1W4XVH5 A0A3G1SUY5 V5GPH2 A0A161MEN8 B4JGS2 A0A1A9VFV9 D6X1R1 A0A1B1UZQ0 D3TSC7 A0A232FGG3 A0A1S6M248 A0A0T6B7V7 A0A069DUZ5 A0A1L8DFJ9 A0A1L1YME0 A0A0N7Z9E6 A0A0V0G3F6 A0A2J7RFL5 T1I122 A0A1I9WLE3 A0A1B0CIV0 A0A023EUA8 A0A182K3I9 A0A182VYQ1 A0A084VXP8 A0A2A3EDR1 V9IKE7 A0A088ADH1 A0A182P806 A0A1A9ZZ34 Q9XYC8 A0A1D1YDC1 J3JVB5 E0VJX6 A0A0U1XFD8 M9TFX8 A0A182XHK3 A0A182MD68
Pubmed
19121390
20158164
28067269
26354079
1387326
23622113
+ More
22651552 22118469 1834020 26108605 24495485 25315136 29908900 25348373 25830018 21347285 20798317 21719571 24508170 30249741 17994087 15632085 23185243 9662471 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26369729 18057021 8940044 12537569 28004739 18362917 19820115 20353571 28648823 26334808 27129103 27538518 24945155 24438588 10212245 17510324 22516182 23537049 20566863 23950915
22651552 22118469 1834020 26108605 24495485 25315136 29908900 25348373 25830018 21347285 20798317 21719571 24508170 30249741 17994087 15632085 23185243 9662471 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 26369729 18057021 8940044 12537569 28004739 18362917 19820115 20353571 28648823 26334808 27129103 27538518 24945155 24438588 10212245 17510324 22516182 23537049 20566863 23950915
EMBL
EF107513
ABK97410.1
BABH01020872
EU727173
ACE78271.1
RSAL01000019
+ More
RVE52771.1 KX685520 AQQ72786.1 KQ459144 KPJ03596.1 X64354 KQ461111 KPJ08932.1 GAIX01003435 JAA89125.1 AK401368 BAM17990.1 AGBW02012914 OWR44221.1 GU370066 ADK94761.1 NWSH01006817 PCG63226.1 S61797 JRES01001160 KNC25029.1 GFDG01001786 JAV17013.1 GAMC01003588 JAC02968.1 KA647427 AFP62056.1 GDHF01009311 JAI43003.1 MH048891 AWW06963.1 KQ982944 KYQ49036.1 GDHF01029014 GDHF01026076 GDHF01018753 GDHF01007591 JAI23300.1 JAI26238.1 JAI33561.1 JAI44723.1 GAKP01014274 GAKP01014273 JAC44678.1 GBXI01010196 GBXI01008895 JAD04096.1 JAD05397.1 KQ977349 KYN03377.1 KQ980673 KYN14604.1 KK855802 PTY23925.1 KQ976433 KYM87346.1 ADTU01027231 GEBQ01029214 JAT10763.1 GL441481 EFN64763.1 KQ981169 KYN45351.1 GECU01032541 JAS75165.1 KQ434803 KZC05953.1 OUUW01000007 SPP82559.1 GL888207 EGI65100.1 CVRI01000012 CRK89474.1 GL449414 EFN82772.1 KK107019 QOIP01000007 EZA62789.1 RLU20950.1 SPP82558.1 GFDL01009738 JAV25307.1 CH479186 EDW39130.1 CM000070 EAL26924.1 KRS99667.1 AF037468 DS231861 AAC04806.1 EDS39442.1 CH940656 EDW58695.1 AE014297 ACZ94894.1 CM000160 EDW97128.1 CP012526 ALC46169.1 CH902623 EDV30327.1 CH964272 EDW83646.1 CH954181 EDV49337.1 KY056637 AQU14352.1 GDAI01001395 JAI16208.1 CH933806 EDW15311.1 KRG01514.1 X67839 AY051623 BT001302 CM000364 EDX13185.1 KQ414581 KOC70995.1 JXJN01007683 CH480815 EDW42428.1 GEZM01064648 JAV68673.1 MG243597 AXU38342.1 GALX01004994 JAB63472.1 GEMB01004297 JAR98975.1 CH916369 EDV92676.1 KQ971371 EFA10717.1 KU550966 ANW09712.1 CCAG010014863 EZ424329 ADD20605.1 NNAY01000282 OXU29519.1 KY056636 AQU14351.1 LJIG01009260 KRT83436.1 GBGD01001243 JAC87646.1 GFDF01008980 JAV05104.1 KM434188 AKM20772.1 GDKW01000763 JAI55832.1 GECL01003505 JAP02619.1 NEVH01004410 PNF39632.1 ACPB03016230 KU932327 APA33963.1 AJWK01013788 GAPW01001097 JAC12501.1 ATLV01018141 KE525214 KFB42742.1 KZ288271 PBC29840.1 JR049481 AEY60976.1 AF092934 CH477360 AAD27666.1 EAT42677.1 GDJX01015304 JAT52632.1 APGK01058793 BT127182 KB741292 KB632334 AEE62144.1 ENN70307.1 ERL92765.1 DS235231 EEB13682.1 KF896233 AIT55914.1 KC473163 AGJ51963.1 AXCM01008181
RVE52771.1 KX685520 AQQ72786.1 KQ459144 KPJ03596.1 X64354 KQ461111 KPJ08932.1 GAIX01003435 JAA89125.1 AK401368 BAM17990.1 AGBW02012914 OWR44221.1 GU370066 ADK94761.1 NWSH01006817 PCG63226.1 S61797 JRES01001160 KNC25029.1 GFDG01001786 JAV17013.1 GAMC01003588 JAC02968.1 KA647427 AFP62056.1 GDHF01009311 JAI43003.1 MH048891 AWW06963.1 KQ982944 KYQ49036.1 GDHF01029014 GDHF01026076 GDHF01018753 GDHF01007591 JAI23300.1 JAI26238.1 JAI33561.1 JAI44723.1 GAKP01014274 GAKP01014273 JAC44678.1 GBXI01010196 GBXI01008895 JAD04096.1 JAD05397.1 KQ977349 KYN03377.1 KQ980673 KYN14604.1 KK855802 PTY23925.1 KQ976433 KYM87346.1 ADTU01027231 GEBQ01029214 JAT10763.1 GL441481 EFN64763.1 KQ981169 KYN45351.1 GECU01032541 JAS75165.1 KQ434803 KZC05953.1 OUUW01000007 SPP82559.1 GL888207 EGI65100.1 CVRI01000012 CRK89474.1 GL449414 EFN82772.1 KK107019 QOIP01000007 EZA62789.1 RLU20950.1 SPP82558.1 GFDL01009738 JAV25307.1 CH479186 EDW39130.1 CM000070 EAL26924.1 KRS99667.1 AF037468 DS231861 AAC04806.1 EDS39442.1 CH940656 EDW58695.1 AE014297 ACZ94894.1 CM000160 EDW97128.1 CP012526 ALC46169.1 CH902623 EDV30327.1 CH964272 EDW83646.1 CH954181 EDV49337.1 KY056637 AQU14352.1 GDAI01001395 JAI16208.1 CH933806 EDW15311.1 KRG01514.1 X67839 AY051623 BT001302 CM000364 EDX13185.1 KQ414581 KOC70995.1 JXJN01007683 CH480815 EDW42428.1 GEZM01064648 JAV68673.1 MG243597 AXU38342.1 GALX01004994 JAB63472.1 GEMB01004297 JAR98975.1 CH916369 EDV92676.1 KQ971371 EFA10717.1 KU550966 ANW09712.1 CCAG010014863 EZ424329 ADD20605.1 NNAY01000282 OXU29519.1 KY056636 AQU14351.1 LJIG01009260 KRT83436.1 GBGD01001243 JAC87646.1 GFDF01008980 JAV05104.1 KM434188 AKM20772.1 GDKW01000763 JAI55832.1 GECL01003505 JAP02619.1 NEVH01004410 PNF39632.1 ACPB03016230 KU932327 APA33963.1 AJWK01013788 GAPW01001097 JAC12501.1 ATLV01018141 KE525214 KFB42742.1 KZ288271 PBC29840.1 JR049481 AEY60976.1 AF092934 CH477360 AAD27666.1 EAT42677.1 GDJX01015304 JAT52632.1 APGK01058793 BT127182 KB741292 KB632334 AEE62144.1 ENN70307.1 ERL92765.1 DS235231 EEB13682.1 KF896233 AIT55914.1 KC473163 AGJ51963.1 AXCM01008181
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
UP000218220
+ More
UP000037069 UP000095300 UP000095301 UP000075809 UP000078542 UP000078492 UP000078540 UP000005205 UP000000311 UP000078541 UP000076502 UP000268350 UP000007755 UP000183832 UP000008237 UP000053097 UP000279307 UP000008744 UP000001819 UP000002320 UP000008792 UP000192221 UP000000803 UP000002282 UP000092553 UP000007801 UP000007798 UP000008711 UP000009192 UP000000304 UP000053825 UP000092443 UP000092460 UP000001292 UP000192223 UP000001070 UP000078200 UP000007266 UP000092444 UP000215335 UP000235965 UP000015103 UP000092461 UP000075881 UP000075920 UP000030765 UP000242457 UP000005203 UP000075885 UP000092445 UP000008820 UP000019118 UP000030742 UP000009046 UP000076407 UP000075883
UP000037069 UP000095300 UP000095301 UP000075809 UP000078542 UP000078492 UP000078540 UP000005205 UP000000311 UP000078541 UP000076502 UP000268350 UP000007755 UP000183832 UP000008237 UP000053097 UP000279307 UP000008744 UP000001819 UP000002320 UP000008792 UP000192221 UP000000803 UP000002282 UP000092553 UP000007801 UP000007798 UP000008711 UP000009192 UP000000304 UP000053825 UP000092443 UP000092460 UP000001292 UP000192223 UP000001070 UP000078200 UP000007266 UP000092444 UP000215335 UP000235965 UP000015103 UP000092461 UP000075881 UP000075920 UP000030765 UP000242457 UP000005203 UP000075885 UP000092445 UP000008820 UP000019118 UP000030742 UP000009046 UP000076407 UP000075883
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A1E4A8
B3VBE3
A0A3S2NP74
A0A1S5XVQ5
A0A194QJG9
P31401
+ More
A0A0N1PFD7 S4PMQ2 I4DJA0 A0A212ERZ3 E7BZ73 A0A2A4IUP6 P31410 A0A0L0C0P8 A0A1I8P324 A0A1L8EEN3 W8BUW3 T1PF56 A0A0K8VVU7 A0A2Z4GWT6 A0A151WMF7 A0A0K8W0S0 A0A034VT14 A0A0A1X2S7 A0A195CSA0 A0A151IZY9 A0A2R7WZ85 A0A195BP22 A0A158NVJ8 A0A1B6KH42 E2AP90 A0A195FY38 A0A1B6HKP9 A0A154P289 A0A3B0K915 F4WLD7 A0A1J1HPL7 E2BNJ3 A0A026X5Q0 A0A3B0KFZ3 A0A1Q3FCM0 B4GN04 Q29BS3 O44427 B4MC74 A0A1W4UPV6 E1JIJ5 B4PPC2 A0A0M3QXN3 B3MST7 B4NHZ6 B3P492 A0A1S6M253 A0A0K8TQ02 B4K7G7 P31409 B4QS89 A0A0L7RJE6 A0A1A9YSX7 A0A1B0B2L4 B4HGV7 A0A1Y1L4S1 A0A1W4XVH5 A0A3G1SUY5 V5GPH2 A0A161MEN8 B4JGS2 A0A1A9VFV9 D6X1R1 A0A1B1UZQ0 D3TSC7 A0A232FGG3 A0A1S6M248 A0A0T6B7V7 A0A069DUZ5 A0A1L8DFJ9 A0A1L1YME0 A0A0N7Z9E6 A0A0V0G3F6 A0A2J7RFL5 T1I122 A0A1I9WLE3 A0A1B0CIV0 A0A023EUA8 A0A182K3I9 A0A182VYQ1 A0A084VXP8 A0A2A3EDR1 V9IKE7 A0A088ADH1 A0A182P806 A0A1A9ZZ34 Q9XYC8 A0A1D1YDC1 J3JVB5 E0VJX6 A0A0U1XFD8 M9TFX8 A0A182XHK3 A0A182MD68
A0A0N1PFD7 S4PMQ2 I4DJA0 A0A212ERZ3 E7BZ73 A0A2A4IUP6 P31410 A0A0L0C0P8 A0A1I8P324 A0A1L8EEN3 W8BUW3 T1PF56 A0A0K8VVU7 A0A2Z4GWT6 A0A151WMF7 A0A0K8W0S0 A0A034VT14 A0A0A1X2S7 A0A195CSA0 A0A151IZY9 A0A2R7WZ85 A0A195BP22 A0A158NVJ8 A0A1B6KH42 E2AP90 A0A195FY38 A0A1B6HKP9 A0A154P289 A0A3B0K915 F4WLD7 A0A1J1HPL7 E2BNJ3 A0A026X5Q0 A0A3B0KFZ3 A0A1Q3FCM0 B4GN04 Q29BS3 O44427 B4MC74 A0A1W4UPV6 E1JIJ5 B4PPC2 A0A0M3QXN3 B3MST7 B4NHZ6 B3P492 A0A1S6M253 A0A0K8TQ02 B4K7G7 P31409 B4QS89 A0A0L7RJE6 A0A1A9YSX7 A0A1B0B2L4 B4HGV7 A0A1Y1L4S1 A0A1W4XVH5 A0A3G1SUY5 V5GPH2 A0A161MEN8 B4JGS2 A0A1A9VFV9 D6X1R1 A0A1B1UZQ0 D3TSC7 A0A232FGG3 A0A1S6M248 A0A0T6B7V7 A0A069DUZ5 A0A1L8DFJ9 A0A1L1YME0 A0A0N7Z9E6 A0A0V0G3F6 A0A2J7RFL5 T1I122 A0A1I9WLE3 A0A1B0CIV0 A0A023EUA8 A0A182K3I9 A0A182VYQ1 A0A084VXP8 A0A2A3EDR1 V9IKE7 A0A088ADH1 A0A182P806 A0A1A9ZZ34 Q9XYC8 A0A1D1YDC1 J3JVB5 E0VJX6 A0A0U1XFD8 M9TFX8 A0A182XHK3 A0A182MD68
PDB
6O7X
E-value=8.18753e-115,
Score=1057
Ontologies
KEGG
PATHWAY
GO
PANTHER
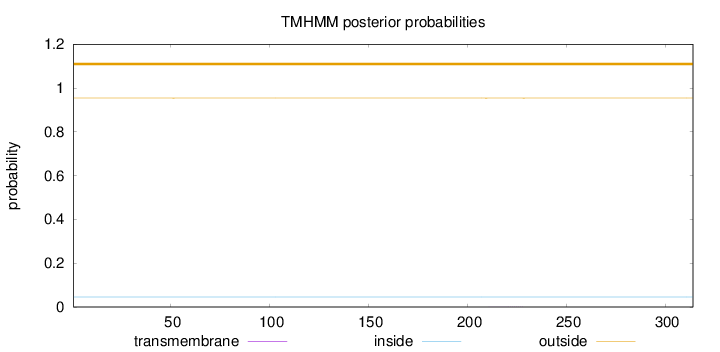
Topology
Length:
314
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00854999999999999
Exp number, first 60 AAs:
0.00077
Total prob of N-in:
0.04586
outside
1 - 314
Population Genetic Test Statistics
Pi
275.081517
Theta
209.111766
Tajima's D
0.8347
CLR
0.702843
CSRT
0.614569271536423
Interpretation
Uncertain
