Gene
KWMTBOMO15750
Pre Gene Modal
BGIBMGA002248
Annotation
PREDICTED:_octopamine_receptor_beta-1R_[Bombyx_mori]
Full name
Octopamine receptor beta-1R
+ More
Octopamine receptor beta-3R
Octopamine receptor beta-3R
Location in the cell
PlasmaMembrane Reliability : 4.951
Sequence
CDS
ATGAAACTGGAAGTCGTATTCAGCCTTGAACTGGGCAACTTCACAGCGGACCTGCAAGATCTATCCACATCCATGTATCTAACGGACACTTACGTGAACAGGACGCTGGAAGTGAATTTGACGACCAATGACTTGGTGTACTCGAATCAGTTCGAAGACGATCCCCTAATTGTGTTTTTAATTATACTAAAATGCATGATAATGCTGTTCATAATATTGGCAGCGATATTTGGCAATCTGTTGGTGATAGTATCGGTGATGAGACACAGAAAATTGCGGGTCATAACGAACTATTTTGTCGTGTCGCTGGCTTTGGCTGATATGTTGGTCGCTATTTGGGCGATGTGCTTCAACTTCAGCGTGGAGATCACGAACGGGGAATGGTTATTTGGCTATTTCATGTGCGACGTTTGGAATTCGTTAGACGTGTACTTCTCGTCGGCGAGCATTCTTCATTTGTGCTGCATCAGTGTCGACCGATACTACGCAATCGTCCAGCCATTGGACTACCCGTTGATTATGACCACGGCCAAGCTCGGCATAATGTTGGCTGTAGTTTGGTGCAGTCCGGCATTGGTCTCGTTTCTACCGATCTTCATGGGTTGGTACACAACCGAGGATCATTTGGACTTCAGAAGACGCTACCCGAAAGTGTGCAGTTTCACAGTCAACAAAGTGTACGCAGTGATATCGAGCAGTATAAGTTTCTGGATTCCCGGTGTGATTATGTTGTTCATGTATTACAGGATTTATGTGGAAGCCGATAGACAGGAGAGAATGCTGTACAGGTAA
Protein
MKLEVVFSLELGNFTADLQDLSTSMYLTDTYVNRTLEVNLTTNDLVYSNQFEDDPLIVFLIILKCMIMLFIILAAIFGNLLVIVSVMRHRKLRVITNYFVVSLALADMLVAIWAMCFNFSVEITNGEWLFGYFMCDVWNSLDVYFSSASILHLCCISVDRYYAIVQPLDYPLIMTTAKLGIMLAVVWCSPALVSFLPIFMGWYTTEDHLDFRRRYPKVCSFTVNKVYAVISSSISFWIPGVIMLFMYYRIYVEADRQERMLYR
Summary
Description
Autoreceptor for octopamine, which is a neurotransmitter, neurohormone, and neuromodulator in invertebrates (PubMed:15816867, PubMed:22553037). Negatively regulates synaptic growth by activating the inhibitory G protein Galphao and limiting cAMP production (PubMed:22553037). Antagonizes the action of Octbeta2R which stimulates synaptic growth (PubMed:22553037).
Autoreceptor for octopamine, which is a neurotransmitter, neurohormone, and neuromodulator in invertebrates (By similarity). Probably also acts as a receptor for tyramine during ecdysone biosynthesis (PubMed:25605909). Required for the biosynthesis of the steroid hormone ecdysone which is necessary for metamorphosis (PubMed:25605909). Involved in activation of prothoracicotropic hormone and insulin-like peptide signaling which is required for the expression of ecdysone biosynthetic genes (PubMed:25605909).
Autoreceptor for octopamine, which is a neurotransmitter, neurohormone, and neuromodulator in invertebrates (By similarity). Probably also acts as a receptor for tyramine during ecdysone biosynthesis (PubMed:25605909). Required for the biosynthesis of the steroid hormone ecdysone which is necessary for metamorphosis (PubMed:25605909). Involved in activation of prothoracicotropic hormone and insulin-like peptide signaling which is required for the expression of ecdysone biosynthetic genes (PubMed:25605909).
Similarity
Belongs to the G-protein coupled receptor 1 family.
Keywords
Alternative splicing
Cell membrane
Complete proteome
G-protein coupled receptor
Glycoprotein
Membrane
Receptor
Reference proteome
Transducer
Transmembrane
Transmembrane helix
Feature
chain Octopamine receptor beta-1R
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9IYB5
A0A2H1W8D0
A0A2A4J6R9
A0A3S2M6T4
T2FFY0
A0A0N1IPC8
+ More
H6V947 A0A0L7L9V2 A0A0L7L9J9 A0A139WBL3 A0A2R7W338 D6WZ39 N1NTW6 X2L2N9 T1HMS5 A0A2J7PYR1 A0A2H4T0P7 A0A067R718 N6TQ91 A0A1B6DVC9 A0A310SLL4 E2BB90 E0VA53 A0A087ZWL9 W0U8S1 E2ATC1 A0A151J0L6 A0A0J7KTY1 A0A0M9A9B9 A0A3L8DR92 A0A0C9QVS2 F4WII1 A0A0C9Q1P1 A0A026VXG0 A0A0L7R2G1 A0A158NZY3 E9J593 A0A1S3DGR0 A0A195BLI6 A0A154P6W6 A0A195F1Z2 A0A195CML9 A0A2H8TLG9 A0A1S3DBJ0 J9K3L5 A0A2S2QQC9 A0A1A9YSY0 A0A1A9ZZ24 A0A1A9VFW2 A0A1I8Q2X1 A0A1B0B2L8 A0A1B0FEL2 E0W0B9 A0A0R1E4D7 A0A0N8P1R6 Q9VCZ3-2 B4K9B5 E1JIT6 B4M0I5 A0A0P9A248 N1NTZ0 B4PMA9 B4JV44 B4R032 B4NJT1 Q9VCZ3 A0A0R1E4B6 B3P8N9 B4HEA3 B3LZZ4 A0A1W4W3K4 A0A0R3NFN9 G7H820 A0A221LCB0 A0A3B0JKB6 G2J627 B4GP31 D6WZ38 Q29BK0 A8E6P6 A0A026VTA1 A0A1Z2RSH0 X2KWS2 A0A0L0BYB7 E9IRG6 A0A1B0GLH6 A0A2J7PTR8 E2C5Q4 Q4LBB6-9 B4M5L3 X2KWM1 A0A067R5V7 A0A158NZY1 A0A3B0JJM1 A0A0L0BYA7 A0A1W4UBG5 B4JGS7 B3P498
H6V947 A0A0L7L9V2 A0A0L7L9J9 A0A139WBL3 A0A2R7W338 D6WZ39 N1NTW6 X2L2N9 T1HMS5 A0A2J7PYR1 A0A2H4T0P7 A0A067R718 N6TQ91 A0A1B6DVC9 A0A310SLL4 E2BB90 E0VA53 A0A087ZWL9 W0U8S1 E2ATC1 A0A151J0L6 A0A0J7KTY1 A0A0M9A9B9 A0A3L8DR92 A0A0C9QVS2 F4WII1 A0A0C9Q1P1 A0A026VXG0 A0A0L7R2G1 A0A158NZY3 E9J593 A0A1S3DGR0 A0A195BLI6 A0A154P6W6 A0A195F1Z2 A0A195CML9 A0A2H8TLG9 A0A1S3DBJ0 J9K3L5 A0A2S2QQC9 A0A1A9YSY0 A0A1A9ZZ24 A0A1A9VFW2 A0A1I8Q2X1 A0A1B0B2L8 A0A1B0FEL2 E0W0B9 A0A0R1E4D7 A0A0N8P1R6 Q9VCZ3-2 B4K9B5 E1JIT6 B4M0I5 A0A0P9A248 N1NTZ0 B4PMA9 B4JV44 B4R032 B4NJT1 Q9VCZ3 A0A0R1E4B6 B3P8N9 B4HEA3 B3LZZ4 A0A1W4W3K4 A0A0R3NFN9 G7H820 A0A221LCB0 A0A3B0JKB6 G2J627 B4GP31 D6WZ38 Q29BK0 A8E6P6 A0A026VTA1 A0A1Z2RSH0 X2KWS2 A0A0L0BYB7 E9IRG6 A0A1B0GLH6 A0A2J7PTR8 E2C5Q4 Q4LBB6-9 B4M5L3 X2KWM1 A0A067R5V7 A0A158NZY1 A0A3B0JJM1 A0A0L0BYA7 A0A1W4UBG5 B4JGS7 B3P498
Pubmed
19121390
26354079
22504014
26227816
18362917
19820115
+ More
18054377 18025266 18316733 20068045 21843505 23604020 24646461 24845553 23537049 20798317 20566863 30249741 21719571 24508170 21347285 21282665 17994087 17550304 15816867 15998303 10731132 12537572 22553037 22303848 12537568 12537573 12537574 16110336 17569856 17569867 15632085 27717101 23185243 26108605 25605909
18054377 18025266 18316733 20068045 21843505 23604020 24646461 24845553 23537049 20798317 20566863 30249741 21719571 24508170 21347285 21282665 17994087 17550304 15816867 15998303 10731132 12537572 22553037 22303848 12537568 12537573 12537574 16110336 17569856 17569867 15632085 27717101 23185243 26108605 25605909
EMBL
BABH01020819
ODYU01006984
SOQ49320.1
NWSH01002989
PCG67140.1
RSAL01000019
+ More
RVE52780.1 KF640249 AGV79326.1 KQ460620 KPJ13529.1 JN815182 AFA55165.1 JTDY01002081 KOB72154.1 KOB72153.1 KQ971372 KYB25290.1 KK854281 PTY14153.1 EFA10771.2 BK005858 DAA64495.1 KJ152558 AHN85841.1 ACPB03014337 NEVH01020342 PNF21440.1 KY352398 ATY68966.1 KK852660 KDR19125.1 APGK01059262 KB741292 KB632186 ENN70476.1 ERL89671.1 GEDC01007661 JAS29637.1 KQ764687 OAD54449.1 GL446948 EFN87040.1 DS235004 EEB10259.1 HF548209 CCO13922.1 GL442548 EFN63288.1 KQ980624 KYN15038.1 LBMM01003252 KMQ93751.1 KQ435721 KOX78519.1 QOIP01000005 RLU22816.1 GBYB01007839 JAG77606.1 GL888175 EGI66112.1 GBYB01007838 GBYB01009473 JAG77605.1 JAG79240.1 KK107816 EZA47549.1 KQ414666 KOC65065.1 ADTU01005126 GL768124 EFZ12018.1 KQ976440 KYM86550.1 KQ434829 KZC07669.1 KQ981856 KYN34595.1 KQ977580 KYN01722.1 GFXV01003045 MBW14850.1 ABLF02038092 ABLF02038101 ABLF02038102 ABLF02038105 GGMS01010734 MBY79937.1 JXJN01007683 CCAG010014863 DS235858 EEB19075.1 CM000160 KRK04060.1 CH902617 KPU80668.1 AJ617526 AJ880687 AJ880688 AE014297 AAF56012.1 CAE84925.1 CAI56428.1 CH933806 EDW15547.1 ACZ94986.1 CH940650 EDW68364.1 KPU80666.1 BK005856 DAA64493.1 EDW98014.1 CH916374 EDV91364.1 CM000364 EDX13938.1 CH964272 EDW85043.1 KRK04061.1 KRK04062.1 CH954182 EDV54134.1 CH480815 EDW43200.1 EDV44184.2 CM000070 KRS99707.1 BT132743 AES58502.1 KX578904 ASM90737.1 OUUW01000005 SPP80772.1 BT128884 AEO17899.1 CH479186 EDW38914.1 EFA10770.2 EAL26997.3 KRS99708.1 BT030838 ABV82220.1 KK107979 EZA47013.1 KX360126 ASA47149.1 KJ152559 AHN85842.1 JRES01001160 KNC25033.1 GL765144 EFZ16826.1 AJWK01035015 AJWK01035016 AJWK01035017 AJWK01035018 AJWK01035019 NEVH01021226 PNF19714.1 GL452786 EFN76724.1 AJ884591 AJ884592 AJ884593 AJ884594 BT029269 CAI56424.1 CH940652 EDW58939.2 KJ152560 AHN85843.1 KK852941 KDR13533.1 ADTU01005104 ADTU01005105 ADTU01005106 OUUW01000007 SPP82554.1 KNC25005.1 CH916369 EDV92681.1 CH954181 EDV49343.1
RVE52780.1 KF640249 AGV79326.1 KQ460620 KPJ13529.1 JN815182 AFA55165.1 JTDY01002081 KOB72154.1 KOB72153.1 KQ971372 KYB25290.1 KK854281 PTY14153.1 EFA10771.2 BK005858 DAA64495.1 KJ152558 AHN85841.1 ACPB03014337 NEVH01020342 PNF21440.1 KY352398 ATY68966.1 KK852660 KDR19125.1 APGK01059262 KB741292 KB632186 ENN70476.1 ERL89671.1 GEDC01007661 JAS29637.1 KQ764687 OAD54449.1 GL446948 EFN87040.1 DS235004 EEB10259.1 HF548209 CCO13922.1 GL442548 EFN63288.1 KQ980624 KYN15038.1 LBMM01003252 KMQ93751.1 KQ435721 KOX78519.1 QOIP01000005 RLU22816.1 GBYB01007839 JAG77606.1 GL888175 EGI66112.1 GBYB01007838 GBYB01009473 JAG77605.1 JAG79240.1 KK107816 EZA47549.1 KQ414666 KOC65065.1 ADTU01005126 GL768124 EFZ12018.1 KQ976440 KYM86550.1 KQ434829 KZC07669.1 KQ981856 KYN34595.1 KQ977580 KYN01722.1 GFXV01003045 MBW14850.1 ABLF02038092 ABLF02038101 ABLF02038102 ABLF02038105 GGMS01010734 MBY79937.1 JXJN01007683 CCAG010014863 DS235858 EEB19075.1 CM000160 KRK04060.1 CH902617 KPU80668.1 AJ617526 AJ880687 AJ880688 AE014297 AAF56012.1 CAE84925.1 CAI56428.1 CH933806 EDW15547.1 ACZ94986.1 CH940650 EDW68364.1 KPU80666.1 BK005856 DAA64493.1 EDW98014.1 CH916374 EDV91364.1 CM000364 EDX13938.1 CH964272 EDW85043.1 KRK04061.1 KRK04062.1 CH954182 EDV54134.1 CH480815 EDW43200.1 EDV44184.2 CM000070 KRS99707.1 BT132743 AES58502.1 KX578904 ASM90737.1 OUUW01000005 SPP80772.1 BT128884 AEO17899.1 CH479186 EDW38914.1 EFA10770.2 EAL26997.3 KRS99708.1 BT030838 ABV82220.1 KK107979 EZA47013.1 KX360126 ASA47149.1 KJ152559 AHN85842.1 JRES01001160 KNC25033.1 GL765144 EFZ16826.1 AJWK01035015 AJWK01035016 AJWK01035017 AJWK01035018 AJWK01035019 NEVH01021226 PNF19714.1 GL452786 EFN76724.1 AJ884591 AJ884592 AJ884593 AJ884594 BT029269 CAI56424.1 CH940652 EDW58939.2 KJ152560 AHN85843.1 KK852941 KDR13533.1 ADTU01005104 ADTU01005105 ADTU01005106 OUUW01000007 SPP82554.1 KNC25005.1 CH916369 EDV92681.1 CH954181 EDV49343.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000037510
UP000007266
+ More
UP000015103 UP000235965 UP000027135 UP000019118 UP000030742 UP000008237 UP000009046 UP000005203 UP000000311 UP000078492 UP000036403 UP000053105 UP000279307 UP000007755 UP000053097 UP000053825 UP000005205 UP000079169 UP000078540 UP000076502 UP000078541 UP000078542 UP000007819 UP000092443 UP000092445 UP000078200 UP000095300 UP000092460 UP000092444 UP000002282 UP000007801 UP000000803 UP000009192 UP000008792 UP000001070 UP000000304 UP000007798 UP000008711 UP000001292 UP000192221 UP000001819 UP000268350 UP000008744 UP000037069 UP000092461
UP000015103 UP000235965 UP000027135 UP000019118 UP000030742 UP000008237 UP000009046 UP000005203 UP000000311 UP000078492 UP000036403 UP000053105 UP000279307 UP000007755 UP000053097 UP000053825 UP000005205 UP000079169 UP000078540 UP000076502 UP000078541 UP000078542 UP000007819 UP000092443 UP000092445 UP000078200 UP000095300 UP000092460 UP000092444 UP000002282 UP000007801 UP000000803 UP000009192 UP000008792 UP000001070 UP000000304 UP000007798 UP000008711 UP000001292 UP000192221 UP000001819 UP000268350 UP000008744 UP000037069 UP000092461
Pfam
PF00001 7tm_1
ProteinModelPortal
H9IYB5
A0A2H1W8D0
A0A2A4J6R9
A0A3S2M6T4
T2FFY0
A0A0N1IPC8
+ More
H6V947 A0A0L7L9V2 A0A0L7L9J9 A0A139WBL3 A0A2R7W338 D6WZ39 N1NTW6 X2L2N9 T1HMS5 A0A2J7PYR1 A0A2H4T0P7 A0A067R718 N6TQ91 A0A1B6DVC9 A0A310SLL4 E2BB90 E0VA53 A0A087ZWL9 W0U8S1 E2ATC1 A0A151J0L6 A0A0J7KTY1 A0A0M9A9B9 A0A3L8DR92 A0A0C9QVS2 F4WII1 A0A0C9Q1P1 A0A026VXG0 A0A0L7R2G1 A0A158NZY3 E9J593 A0A1S3DGR0 A0A195BLI6 A0A154P6W6 A0A195F1Z2 A0A195CML9 A0A2H8TLG9 A0A1S3DBJ0 J9K3L5 A0A2S2QQC9 A0A1A9YSY0 A0A1A9ZZ24 A0A1A9VFW2 A0A1I8Q2X1 A0A1B0B2L8 A0A1B0FEL2 E0W0B9 A0A0R1E4D7 A0A0N8P1R6 Q9VCZ3-2 B4K9B5 E1JIT6 B4M0I5 A0A0P9A248 N1NTZ0 B4PMA9 B4JV44 B4R032 B4NJT1 Q9VCZ3 A0A0R1E4B6 B3P8N9 B4HEA3 B3LZZ4 A0A1W4W3K4 A0A0R3NFN9 G7H820 A0A221LCB0 A0A3B0JKB6 G2J627 B4GP31 D6WZ38 Q29BK0 A8E6P6 A0A026VTA1 A0A1Z2RSH0 X2KWS2 A0A0L0BYB7 E9IRG6 A0A1B0GLH6 A0A2J7PTR8 E2C5Q4 Q4LBB6-9 B4M5L3 X2KWM1 A0A067R5V7 A0A158NZY1 A0A3B0JJM1 A0A0L0BYA7 A0A1W4UBG5 B4JGS7 B3P498
H6V947 A0A0L7L9V2 A0A0L7L9J9 A0A139WBL3 A0A2R7W338 D6WZ39 N1NTW6 X2L2N9 T1HMS5 A0A2J7PYR1 A0A2H4T0P7 A0A067R718 N6TQ91 A0A1B6DVC9 A0A310SLL4 E2BB90 E0VA53 A0A087ZWL9 W0U8S1 E2ATC1 A0A151J0L6 A0A0J7KTY1 A0A0M9A9B9 A0A3L8DR92 A0A0C9QVS2 F4WII1 A0A0C9Q1P1 A0A026VXG0 A0A0L7R2G1 A0A158NZY3 E9J593 A0A1S3DGR0 A0A195BLI6 A0A154P6W6 A0A195F1Z2 A0A195CML9 A0A2H8TLG9 A0A1S3DBJ0 J9K3L5 A0A2S2QQC9 A0A1A9YSY0 A0A1A9ZZ24 A0A1A9VFW2 A0A1I8Q2X1 A0A1B0B2L8 A0A1B0FEL2 E0W0B9 A0A0R1E4D7 A0A0N8P1R6 Q9VCZ3-2 B4K9B5 E1JIT6 B4M0I5 A0A0P9A248 N1NTZ0 B4PMA9 B4JV44 B4R032 B4NJT1 Q9VCZ3 A0A0R1E4B6 B3P8N9 B4HEA3 B3LZZ4 A0A1W4W3K4 A0A0R3NFN9 G7H820 A0A221LCB0 A0A3B0JKB6 G2J627 B4GP31 D6WZ38 Q29BK0 A8E6P6 A0A026VTA1 A0A1Z2RSH0 X2KWS2 A0A0L0BYB7 E9IRG6 A0A1B0GLH6 A0A2J7PTR8 E2C5Q4 Q4LBB6-9 B4M5L3 X2KWM1 A0A067R5V7 A0A158NZY1 A0A3B0JJM1 A0A0L0BYA7 A0A1W4UBG5 B4JGS7 B3P498
PDB
6IBL
E-value=8.58728e-40,
Score=409
Ontologies
GO
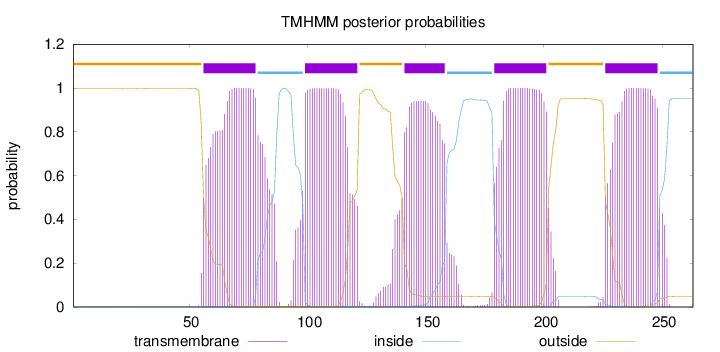
Topology
Subcellular location
Cell membrane
Length:
263
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
113.32138
Exp number, first 60 AAs:
3.53533
Total prob of N-in:
0.00368
outside
1 - 55
TMhelix
56 - 78
inside
79 - 98
TMhelix
99 - 121
outside
122 - 140
TMhelix
141 - 158
inside
159 - 178
TMhelix
179 - 201
outside
202 - 225
TMhelix
226 - 248
inside
249 - 263
Population Genetic Test Statistics
Pi
193.510363
Theta
153.633244
Tajima's D
0.781141
CLR
0.604688
CSRT
0.592170391480426
Interpretation
Uncertain
