Pre Gene Modal
BGIBMGA002330
Annotation
proline_synthetase_co-transcribed_bacterial-like_protein_[Bombyx_mori]
Full name
Pyridoxal phosphate homeostasis protein
Alternative Name
Proline synthase co-transcribed bacterial homolog protein
Location in the cell
Cytoplasmic Reliability : 1.681 Mitochondrial Reliability : 1.723
Sequence
CDS
ATGAGGATAATTAAATCTTCAGTTATGACCAGTGAAGTCGATGCTAAAGTTGACATAATGTATGGATTGAAGACGGTTCTTTCACAAATTGAAATTGCCGTGGCCAGAAGAAGTAAAGACCTACCTCAGATAGCACCAAGATTGGTTGCTGTATCAAAAATTAAACCTGTCGAACTTATTGTGGAAGCTTACAATGCTGGCCAGAGGCACTTCGGAGAAAACTATGTAAACGAATTAAGTGATAAAGCCAGTGATCCATTAATACTGGAAAAATGCAAAGACATTAAATGGCACTTCATTGGACATTTACAAACAAACAAGATTAACAAACTACTTGGTTCGCCAGGCTTGTTTATGGTTGAAACAGTTGATTCAGAAAAATTAGCTGATAATTTGAATAAGCAATGGTTGAAATATAGGAAGGAGAAGGAAAGATTGAGAGTTATGGTACAAGTTAATACTAGTGGTGAACAAGCAAAAAGCGGCTTAGAACCATTGGAAACAACAAAAGCTGTTGAGCACATTCTCGAAAACTGTCCAAACCTAGATTTCCAGGGACTGATGACCATAGGGCAGTATGACTATGATATTACCAAAGGACCAAATCCTGACTATCTGTGCCTAATAAAATGCAGGCAAGAAGTGTGTGAGAAGTTGAATTTGGATATCAAAGATGTTGAGTTATCAATGGGAATGTCTTCAGATTTTGCACATGCTATTGAATTAGGCGCAACTACAGTTCGGGTTGGTTCTACTATTTTTGGAGCAAGGCCGCCGAAAAATTCATAA
Protein
MRIIKSSVMTSEVDAKVDIMYGLKTVLSQIEIAVARRSKDLPQIAPRLVAVSKIKPVELIVEAYNAGQRHFGENYVNELSDKASDPLILEKCKDIKWHFIGHLQTNKINKLLGSPGLFMVETVDSEKLADNLNKQWLKYRKEKERLRVMVQVNTSGEQAKSGLEPLETTKAVEHILENCPNLDFQGLMTIGQYDYDITKGPNPDYLCLIKCRQEVCEKLNLDIKDVELSMGMSSDFAHAIELGATTVRVGSTIFGARPPKNS
Summary
Description
Pyridoxal 5'-phosphate (PLP)-binding protein, which may be involved in intracellular homeostatic regulation of pyridoxal 5'-phosphate (PLP), the active form of vitamin B6.
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the pyridoxal phosphate-binding protein YggS/PROSC family.
Feature
chain Pyridoxal phosphate homeostasis protein
Uniprot
Q2F662
H9IYJ6
A0A0N1IF25
A0A194QIG2
A0A2W1BFV7
A0A2H1WHM4
+ More
A0A212EL27 S4PA59 A0A1B6LK24 N6U5X1 A0A3Q2QN12 A0A3B3XMK1 U4U5Z1 A0A146NLZ2 M3ZQ71 A0A3P8TF67 A0A3Q1ETS8 A0A096M0T1 A0A3Q1AQU9 A0A3B4Y9B0 A0A3B4ULT2 A0A087YE34 A0A3B3TVZ9 A0A3P9NEC9 A0A3Q0RHT6 A0A3B5KNR0 A0A3Q2Y2Z8 A0A1A8GCQ5 A0A1B6KJU9 A0A1Y1K2I7 A0A3B4ULV7 A0A3B4D0E5 A0A3B5B705 A0A3Q3A941 I3IVL2 A0A3B4AFA8 A0A1A8I7S6 A0A1A8QEY3 A0A3B4FQB2 A0A3Q2V3P0 A0A3Q4MIC5 A0A3P8NWT4 A0A3P9D6M4 A0A1A8SHR4 A0A1A8D5X9 A0A0N8JZF4 A0A3Q2E3Z2 A0A1A8RGZ5 A0A3Q1JRS6 A0A1A7WSE9 A0A3B3T2C1 A0A0S7JX53 A0A3B3BVM8 A0A3Q3IF63 H3CCX0 A0A3S2PUW4 A0A3Q3MFD8 I3IVL3 A0A3B1ID89 A0A3Q7Q8B4 H2S9R8 A0A158N9P3 A0A088AER7 H9G3C9 W5U6X0 A0A1B6JE58 H0Z2K7 D6WZU8 A0A1B6HFI3 A0A2I4BVL3 A0A315WD19 A0A2A3ECU6 A0A1W7REK3 B5X4B5 H2L9W8 A0A3P9LFM9 T1DLS6 A0A218UK76 C1BF86 H2L9W9 A0A3P9MMP2 J3S9B4 A0A1U7QI70 A0A3Q3FPY7 A0A1V4KHI1 A0A2U9C1U3 A0A2D4FFA1 E1C516 A0A2P4T2D1 A0A2U3VI01 A0A195F7Q6 F6TK15 A0A337S203 L5LS66 C1BH33 A0A091HRT6 G1P9A7 F4WTT5 B1H1M3 I3M684
A0A212EL27 S4PA59 A0A1B6LK24 N6U5X1 A0A3Q2QN12 A0A3B3XMK1 U4U5Z1 A0A146NLZ2 M3ZQ71 A0A3P8TF67 A0A3Q1ETS8 A0A096M0T1 A0A3Q1AQU9 A0A3B4Y9B0 A0A3B4ULT2 A0A087YE34 A0A3B3TVZ9 A0A3P9NEC9 A0A3Q0RHT6 A0A3B5KNR0 A0A3Q2Y2Z8 A0A1A8GCQ5 A0A1B6KJU9 A0A1Y1K2I7 A0A3B4ULV7 A0A3B4D0E5 A0A3B5B705 A0A3Q3A941 I3IVL2 A0A3B4AFA8 A0A1A8I7S6 A0A1A8QEY3 A0A3B4FQB2 A0A3Q2V3P0 A0A3Q4MIC5 A0A3P8NWT4 A0A3P9D6M4 A0A1A8SHR4 A0A1A8D5X9 A0A0N8JZF4 A0A3Q2E3Z2 A0A1A8RGZ5 A0A3Q1JRS6 A0A1A7WSE9 A0A3B3T2C1 A0A0S7JX53 A0A3B3BVM8 A0A3Q3IF63 H3CCX0 A0A3S2PUW4 A0A3Q3MFD8 I3IVL3 A0A3B1ID89 A0A3Q7Q8B4 H2S9R8 A0A158N9P3 A0A088AER7 H9G3C9 W5U6X0 A0A1B6JE58 H0Z2K7 D6WZU8 A0A1B6HFI3 A0A2I4BVL3 A0A315WD19 A0A2A3ECU6 A0A1W7REK3 B5X4B5 H2L9W8 A0A3P9LFM9 T1DLS6 A0A218UK76 C1BF86 H2L9W9 A0A3P9MMP2 J3S9B4 A0A1U7QI70 A0A3Q3FPY7 A0A1V4KHI1 A0A2U9C1U3 A0A2D4FFA1 E1C516 A0A2P4T2D1 A0A2U3VI01 A0A195F7Q6 F6TK15 A0A337S203 L5LS66 C1BH33 A0A091HRT6 G1P9A7 F4WTT5 B1H1M3 I3M684
Pubmed
19121390
26354079
28756777
22118469
23622113
23537049
+ More
23542700 28004739 25186727 25463417 29240929 29451363 15496914 25329095 21551351 21347285 21881562 23127152 20360741 18362917 19820115 29703783 26358130 20433749 17554307 23758969 23025625 15592404 20431018 17975172 21993624 21719571 23594743
23542700 28004739 25186727 25463417 29240929 29451363 15496914 25329095 21551351 21347285 21881562 23127152 20360741 18362917 19820115 29703783 26358130 20433749 17554307 23758969 23025625 15592404 20431018 17975172 21993624 21719571 23594743
EMBL
DQ311210
ABD36155.1
BABH01020804
KQ460794
KPJ12185.1
KQ458761
+ More
KPJ05184.1 KZ150122 PZC73161.1 ODYU01008638 SOQ52392.1 AGBW02014148 OWR42185.1 GAIX01006532 JAA86028.1 GEBQ01015980 JAT23997.1 APGK01038535 KB740960 ENN77010.1 KB631679 ERL85350.1 GCES01153895 JAQ32427.1 AYCK01013129 HAEB01014345 HAEC01000072 SBQ68149.1 GEBQ01028248 JAT11729.1 GEZM01097336 JAV54270.1 AERX01047889 HAED01006905 SBQ93052.1 HAEF01013247 HAEG01012198 SBR91719.1 HAEI01014745 SBS17214.1 HADZ01022295 HAEA01000409 SBP86236.1 JARO02004087 KPP69185.1 HAEH01016913 SBS05301.1 HADW01007240 HADX01008700 SBP08640.1 GBYX01228849 GBYX01228845 GBYX01228844 GBYX01228842 GBYX01228840 JAO68987.1 CM012443 RVE70290.1 ADTU01009726 AAWZ02027240 JT405720 AHH37294.1 GECU01010318 JAS97388.1 ABQF01056100 ABQF01056101 KQ971372 EFA09643.1 GECU01034347 JAS73359.1 NHOQ01000023 PWA33756.1 KZ288301 PBC28841.1 GDAY02001870 JAV49562.1 BT045884 ACI34146.1 GAAZ01001892 JAA96051.1 MUZQ01000263 OWK53830.1 BT073265 ACO07689.1 JU175494 GBEX01002615 AFJ51018.1 JAI11945.1 LSYS01003090 OPJ83909.1 CP026253 AWP09102.1 IACJ01069160 LAA46184.1 AADN05000189 PPHD01011511 POI30519.1 KQ981727 KYN36650.1 AAMC01017469 AAMC01017470 AAMC01017471 AANG04001605 KB108646 ELK28865.1 BT073912 ACO08336.1 KL217810 KFO99003.1 AAPE02012457 AAPE02012458 GL888344 EGI62400.1 CT025912 BC160662 AAI60662.1 AGTP01097604 AGTP01097605 AGTP01097606
KPJ05184.1 KZ150122 PZC73161.1 ODYU01008638 SOQ52392.1 AGBW02014148 OWR42185.1 GAIX01006532 JAA86028.1 GEBQ01015980 JAT23997.1 APGK01038535 KB740960 ENN77010.1 KB631679 ERL85350.1 GCES01153895 JAQ32427.1 AYCK01013129 HAEB01014345 HAEC01000072 SBQ68149.1 GEBQ01028248 JAT11729.1 GEZM01097336 JAV54270.1 AERX01047889 HAED01006905 SBQ93052.1 HAEF01013247 HAEG01012198 SBR91719.1 HAEI01014745 SBS17214.1 HADZ01022295 HAEA01000409 SBP86236.1 JARO02004087 KPP69185.1 HAEH01016913 SBS05301.1 HADW01007240 HADX01008700 SBP08640.1 GBYX01228849 GBYX01228845 GBYX01228844 GBYX01228842 GBYX01228840 JAO68987.1 CM012443 RVE70290.1 ADTU01009726 AAWZ02027240 JT405720 AHH37294.1 GECU01010318 JAS97388.1 ABQF01056100 ABQF01056101 KQ971372 EFA09643.1 GECU01034347 JAS73359.1 NHOQ01000023 PWA33756.1 KZ288301 PBC28841.1 GDAY02001870 JAV49562.1 BT045884 ACI34146.1 GAAZ01001892 JAA96051.1 MUZQ01000263 OWK53830.1 BT073265 ACO07689.1 JU175494 GBEX01002615 AFJ51018.1 JAI11945.1 LSYS01003090 OPJ83909.1 CP026253 AWP09102.1 IACJ01069160 LAA46184.1 AADN05000189 PPHD01011511 POI30519.1 KQ981727 KYN36650.1 AAMC01017469 AAMC01017470 AAMC01017471 AANG04001605 KB108646 ELK28865.1 BT073912 ACO08336.1 KL217810 KFO99003.1 AAPE02012457 AAPE02012458 GL888344 EGI62400.1 CT025912 BC160662 AAI60662.1 AGTP01097604 AGTP01097605 AGTP01097606
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000019118
UP000265000
+ More
UP000261480 UP000030742 UP000002852 UP000265080 UP000257200 UP000028760 UP000257160 UP000261360 UP000261420 UP000261500 UP000242638 UP000261340 UP000261380 UP000264820 UP000261440 UP000261400 UP000264800 UP000005207 UP000261520 UP000261460 UP000264840 UP000261580 UP000265100 UP000265160 UP000034805 UP000192224 UP000265020 UP000265040 UP000261540 UP000261560 UP000261600 UP000007303 UP000261640 UP000018467 UP000286641 UP000005226 UP000005205 UP000005203 UP000001646 UP000221080 UP000007754 UP000007266 UP000192220 UP000242457 UP000087266 UP000001038 UP000265180 UP000265200 UP000197619 UP000189706 UP000261660 UP000190648 UP000246464 UP000000539 UP000245340 UP000078541 UP000008143 UP000011712 UP000054308 UP000001074 UP000007755 UP000000437 UP000005215
UP000261480 UP000030742 UP000002852 UP000265080 UP000257200 UP000028760 UP000257160 UP000261360 UP000261420 UP000261500 UP000242638 UP000261340 UP000261380 UP000264820 UP000261440 UP000261400 UP000264800 UP000005207 UP000261520 UP000261460 UP000264840 UP000261580 UP000265100 UP000265160 UP000034805 UP000192224 UP000265020 UP000265040 UP000261540 UP000261560 UP000261600 UP000007303 UP000261640 UP000018467 UP000286641 UP000005226 UP000005205 UP000005203 UP000001646 UP000221080 UP000007754 UP000007266 UP000192220 UP000242457 UP000087266 UP000001038 UP000265180 UP000265200 UP000197619 UP000189706 UP000261660 UP000190648 UP000246464 UP000000539 UP000245340 UP000078541 UP000008143 UP000011712 UP000054308 UP000001074 UP000007755 UP000000437 UP000005215
Pfam
PF01168 Ala_racemase_N
Interpro
SUPFAM
SSF51419
SSF51419
Gene 3D
ProteinModelPortal
Q2F662
H9IYJ6
A0A0N1IF25
A0A194QIG2
A0A2W1BFV7
A0A2H1WHM4
+ More
A0A212EL27 S4PA59 A0A1B6LK24 N6U5X1 A0A3Q2QN12 A0A3B3XMK1 U4U5Z1 A0A146NLZ2 M3ZQ71 A0A3P8TF67 A0A3Q1ETS8 A0A096M0T1 A0A3Q1AQU9 A0A3B4Y9B0 A0A3B4ULT2 A0A087YE34 A0A3B3TVZ9 A0A3P9NEC9 A0A3Q0RHT6 A0A3B5KNR0 A0A3Q2Y2Z8 A0A1A8GCQ5 A0A1B6KJU9 A0A1Y1K2I7 A0A3B4ULV7 A0A3B4D0E5 A0A3B5B705 A0A3Q3A941 I3IVL2 A0A3B4AFA8 A0A1A8I7S6 A0A1A8QEY3 A0A3B4FQB2 A0A3Q2V3P0 A0A3Q4MIC5 A0A3P8NWT4 A0A3P9D6M4 A0A1A8SHR4 A0A1A8D5X9 A0A0N8JZF4 A0A3Q2E3Z2 A0A1A8RGZ5 A0A3Q1JRS6 A0A1A7WSE9 A0A3B3T2C1 A0A0S7JX53 A0A3B3BVM8 A0A3Q3IF63 H3CCX0 A0A3S2PUW4 A0A3Q3MFD8 I3IVL3 A0A3B1ID89 A0A3Q7Q8B4 H2S9R8 A0A158N9P3 A0A088AER7 H9G3C9 W5U6X0 A0A1B6JE58 H0Z2K7 D6WZU8 A0A1B6HFI3 A0A2I4BVL3 A0A315WD19 A0A2A3ECU6 A0A1W7REK3 B5X4B5 H2L9W8 A0A3P9LFM9 T1DLS6 A0A218UK76 C1BF86 H2L9W9 A0A3P9MMP2 J3S9B4 A0A1U7QI70 A0A3Q3FPY7 A0A1V4KHI1 A0A2U9C1U3 A0A2D4FFA1 E1C516 A0A2P4T2D1 A0A2U3VI01 A0A195F7Q6 F6TK15 A0A337S203 L5LS66 C1BH33 A0A091HRT6 G1P9A7 F4WTT5 B1H1M3 I3M684
A0A212EL27 S4PA59 A0A1B6LK24 N6U5X1 A0A3Q2QN12 A0A3B3XMK1 U4U5Z1 A0A146NLZ2 M3ZQ71 A0A3P8TF67 A0A3Q1ETS8 A0A096M0T1 A0A3Q1AQU9 A0A3B4Y9B0 A0A3B4ULT2 A0A087YE34 A0A3B3TVZ9 A0A3P9NEC9 A0A3Q0RHT6 A0A3B5KNR0 A0A3Q2Y2Z8 A0A1A8GCQ5 A0A1B6KJU9 A0A1Y1K2I7 A0A3B4ULV7 A0A3B4D0E5 A0A3B5B705 A0A3Q3A941 I3IVL2 A0A3B4AFA8 A0A1A8I7S6 A0A1A8QEY3 A0A3B4FQB2 A0A3Q2V3P0 A0A3Q4MIC5 A0A3P8NWT4 A0A3P9D6M4 A0A1A8SHR4 A0A1A8D5X9 A0A0N8JZF4 A0A3Q2E3Z2 A0A1A8RGZ5 A0A3Q1JRS6 A0A1A7WSE9 A0A3B3T2C1 A0A0S7JX53 A0A3B3BVM8 A0A3Q3IF63 H3CCX0 A0A3S2PUW4 A0A3Q3MFD8 I3IVL3 A0A3B1ID89 A0A3Q7Q8B4 H2S9R8 A0A158N9P3 A0A088AER7 H9G3C9 W5U6X0 A0A1B6JE58 H0Z2K7 D6WZU8 A0A1B6HFI3 A0A2I4BVL3 A0A315WD19 A0A2A3ECU6 A0A1W7REK3 B5X4B5 H2L9W8 A0A3P9LFM9 T1DLS6 A0A218UK76 C1BF86 H2L9W9 A0A3P9MMP2 J3S9B4 A0A1U7QI70 A0A3Q3FPY7 A0A1V4KHI1 A0A2U9C1U3 A0A2D4FFA1 E1C516 A0A2P4T2D1 A0A2U3VI01 A0A195F7Q6 F6TK15 A0A337S203 L5LS66 C1BH33 A0A091HRT6 G1P9A7 F4WTT5 B1H1M3 I3M684
PDB
1CT5
E-value=6.54075e-36,
Score=375
Ontologies
PANTHER
Topology
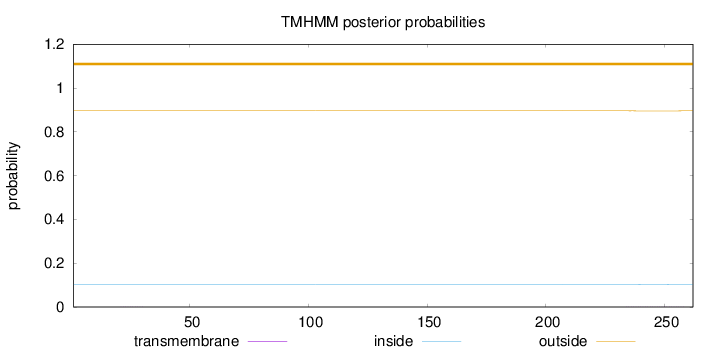
Length:
262
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01975
Exp number, first 60 AAs:
0.0024
Total prob of N-in:
0.10394
outside
1 - 262
Population Genetic Test Statistics
Pi
236.664261
Theta
150.431033
Tajima's D
1.768103
CLR
17.77153
CSRT
0.842157892105395
Interpretation
Uncertain
